Meta-analysis derived (MAD) transcriptome of psoriasis defines the "core" pathogenesis of disease
- PMID: 22957057
- PMCID: PMC3434204
- DOI: 10.1371/journal.pone.0044274
Meta-analysis derived (MAD) transcriptome of psoriasis defines the "core" pathogenesis of disease
Abstract
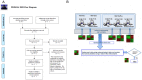
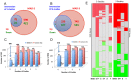
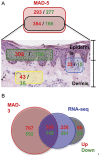
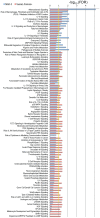
The cause of psoriasis, a common chronic inflammatory skin disease, is not fully understood. Microarray experiments have been widely used in recent years to identify genes associated with psoriasis pathology, by comparing expression levels of lesional (LS) with adjacent non-lesional (NL) skin. It is commonly observed that the differentially expressed genes (DEGs) differ greatly across experiments, due to variations introduced in the microarray experiment pipeline. Therefore, a statistically based meta-analytic approach, which combines the results of individual studies, is warranted. In this study, a meta-analysis was conducted on 5 microarray data sets, including 193 LS and NL pairs. We termed this the Meta-Analysis Derived (MAD) transcriptome. In "MAD-5" transcriptome, 677 genes were up-regulated and 443 were down-regulated in LS skin compared to NL skin. This represents a much larger set than the intersection of DEGs of these 5 studies, which consisted of 100 DEGs. We also analyzed 3 of the studies conducted on the Affymetrix hgu133plus2 chips and found a greater number of DEGs (1084 up- and 748 down-regulated). Top canonical pathways over-represented in the MAD transcriptome include Atherosclerosis Signaling and Fatty Acid Metabolism, while several "new" genes identified are involved in Cardiovascular Development and Lipid Metabolism. These findings highlight the relationship between psoriasis and systemic manifestations such as the metabolic syndrome and cardiovascular disease. Then, the Meta Threshold Gradient Descent Regularization (MTGDR) algorithm was used to select potential markers distinguishing LS and NL skin. The resulting set (20 genes) contained many genes that were part of the residual disease genomic profile (RDGP) or "molecular scar" after successful treatment, and also genes subject to differential methylation in LS tissues. To conclude, this MAD transcriptome yielded a reference list of reliable psoriasis DEGs, and represents a robust pool of candidates for further discovery of pathogenesis and treatment evaluation.
Conflict of interest statement
Figures
References
-
- Bowcock AM, Shannon W, Du F, Duncan J, Cao K, et al. (2001) Insights into psoriasis and other inflammatory diseases from large-scale gene expression studies. Hum Mol Genet 10: 1793–1805. - PubMed
-
- Oestreicher JL, Walters IB, Kikuchi T, Gilleaudeau P, Surette J, et al. (2001) Molecular classification of psoriasis disease-associated genes through pharmacogenomic expression profiling. Pharmacogenomics J 1: 272–287. - PubMed
-
- Zhou X, Krueger JG, Kao MC, Lee E, Du F, et al. (2003) Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics 13: 69–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

