Agnathan VIP, PACAP and their receptors: ancestral origins of today's highly diversified forms
- PMID: 22957100
- PMCID: PMC3434177
- DOI: 10.1371/journal.pone.0044691
Agnathan VIP, PACAP and their receptors: ancestral origins of today's highly diversified forms
Abstract
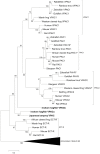
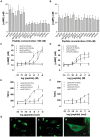
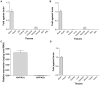
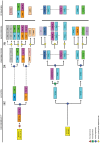
VIP and PACAP are pleiotropic peptides belonging to the secretin superfamily of brain-gut peptides and interact specifically with three receptors (VPAC(1), PAC(1) and VPAC(2)) from the class II B G protein-coupled receptor family. There is immense interest regarding their molecular evolution which is often described closely alongside gene and/or genome duplications. Despite the wide array of information available in various vertebrates and one invertebrate the tunicate, their evolutionary origins remain unresolved. Through searches of genome databases and molecular cloning techniques, the first lamprey VIP/PACAP ligands and VPAC receptors are identified from the Japanese lamprey. In addition, two VPAC receptors (VPACa/b) are identified from inshore hagfish and ligands predicted for sea lamprey. Phylogenetic analyses group these molecules into their respective PHI/VIP, PRP/PACAP and VPAC receptor families and show they resemble ancestral forms. Japanese lamprey VIP/PACAP peptides synthesized were tested with the hagfish VPAC receptors. hfVPACa transduces signal via both adenylyl cylase and phospholipase C pathways, whilst hfVPACb was only able to transduce through the calcium pathway. In contrast to the widespread distribution of VIP/PACAP ligands and receptors in many species, the agnathan PACAP and VPAC receptors were found almost exclusively in the brain. In situ hybridisation further showed their abundance throughout the brain. The range of VIP/PACAP ligands and receptors found are highly useful, providing a glimpse into the evolutionary events both at the structural and functional levels. Though representative of ancestral forms, the VIP/PACAP ligands in particular have retained high sequence conservation indicating the importance of their functions even early in vertebrate evolution. During these nascent stages, only two VPAC receptors are likely responsible for eliciting functions before evolving later into specific subtypes post-Agnatha. We also propose VIP and PACAP's first functions to predominate in the brain, evolving alongside the central nervous system, subsequently establishing peripheral functions.
Conflict of interest statement
Figures



Similar articles
-
Structural and functional identification of the pituitary adenylate cyclase-activating polypeptide receptor VPAC2 from the frog Rana tigrina rugulosa.J Mol Endocrinol. 2001 Oct;27(2):229-38. doi: 10.1677/jme.0.0270229. J Mol Endocrinol. 2001. PMID: 11564605
-
Pituitary adenylate cyclase-activating polypeptide, vasoactive intestinal polypeptide and their receptors: distribution and involvement in the secretion of Podarcis sicula adrenal gland.J Endocrinol. 2008 Feb;196(2):291-303. doi: 10.1677/JOE-07-0127. J Endocrinol. 2008. PMID: 18252952
-
Pharmacology and functions of receptors for vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide: IUPHAR review 1.Br J Pharmacol. 2012 May;166(1):4-17. doi: 10.1111/j.1476-5381.2012.01871.x. Br J Pharmacol. 2012. PMID: 22289055 Free PMC article. Review.
-
Expression and function of vasoactive intestinal peptide, pituitary adenylate cyclase-activating polypeptide, and their receptors in the human adrenal gland.J Clin Endocrinol Metab. 2002 Jun;87(6):2575-80. doi: 10.1210/jcem.87.6.8571. J Clin Endocrinol Metab. 2002. PMID: 12050216
-
Endogenous ligands of PACAP/VIP receptors in the autocrine-paracrine regulation of the adrenal gland.Int Rev Cytol. 2006;249:1-51. doi: 10.1016/S0074-7696(06)49001-X. Int Rev Cytol. 2006. PMID: 16697281 Review.
Cited by
-
Nematode and arthropod genomes provide new insights into the evolution of class 2 B1 GPCRs.PLoS One. 2014 Mar 20;9(3):e92220. doi: 10.1371/journal.pone.0092220. eCollection 2014. PLoS One. 2014. PMID: 24651821 Free PMC article.
-
A Molecular Dynamics Study of Vasoactive Intestinal Peptide Receptor 1 and the Basis of Its Therapeutic Antagonism.Int J Mol Sci. 2019 Sep 5;20(18):4348. doi: 10.3390/ijms20184348. Int J Mol Sci. 2019. PMID: 31491880 Free PMC article.
-
Functional Pairing of Class B1 Ligand-GPCR in Cephalochordate Provides Evidence of the Origin of PTH and PACAP/Glucagon Receptor Family.Mol Biol Evol. 2015 Aug;32(8):2048-59. doi: 10.1093/molbev/msv087. Epub 2015 Apr 3. Mol Biol Evol. 2015. PMID: 25841489 Free PMC article.
-
Gene Expression Analysis Links Autocrine Vasoactive Intestinal Peptide and ZEB1 in Gastrointestinal Cancers.Cancers (Basel). 2023 Jun 22;15(13):3284. doi: 10.3390/cancers15133284. Cancers (Basel). 2023. PMID: 37444395 Free PMC article.
-
PACAP deficiency as a model of aging.Geroscience. 2018 Dec;40(5-6):437-452. doi: 10.1007/s11357-018-0045-8. Epub 2018 Oct 22. Geroscience. 2018. PMID: 30345481 Free PMC article. Review.
References
-
- Said SI, Mutt V (1970) Polypeptide with broad biological activity: isolation from small intestine. Science 169: 1217–1218. - PubMed
-
- Miyata A, Arimura A, Dahl RR, Minamino N, Uehara A, et al. (1989) Isolation of a novel 38 residue-hypothalamic polypeptide which stimulates adenylate cyclase in pituitary cells. Biochem Biophys Res Commun 164: 567–574. - PubMed
-
- Ishihara T, Shigemoto R, Mori K, Takahashi K, Nagata S (1992) Functional expression and tissue distribution of a novel receptor for vasoactive intestinal polypeptide. Neuron 8: 811–819. - PubMed
-
- Lutz EM, Sheward WJ, West KM, Morrow JA, Fink G, et al. (1993) The VIP2 receptor: molecular characterisation of a cDNA encoding a novel receptor for vasoactive intestinal peptide. FEBS Lett 334: 3–8. - PubMed
-
- Hashimoto H, Ishihara T, Shigemoto R, Mori K, Nagata S (1993) Molecular cloning and tissue distribution of a receptor for pituitary adenylate cyclase-activating polypeptide. Neuron 11: 333–342. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

