Targeting the cancer cell cycle by cold atmospheric plasma
- PMID: 22957140
- PMCID: PMC3434394
- DOI: 10.1038/srep00636
Targeting the cancer cell cycle by cold atmospheric plasma
Abstract
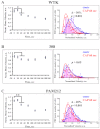
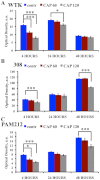
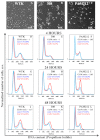
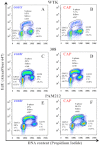
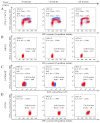
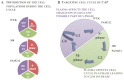
Cold atmospheric plasma (CAP), a technology based on quasi-neutral ionized gas at low temperatures, is currently being evaluated as a new highly selective alternative addition to existing cancer therapies. Here, we present a first attempt to identify the mechanism of CAP action. CAP induced a robust ~2-fold G2/M increase in two different types of cancer cells with different degrees of tumorigenicity. We hypothesize that the increased sensitivity of cancer cells to CAP treatment is caused by differences in the distribution of cancer cells and normal cells within the cell cycle. The expression of γH2A.X (pSer139), an oxidative stress reporter indicating S-phase damage, is enhanced specifically within CAP treated cells in the S phase of the cell cycle. Together with a significant decrease in EdU-incorporation after CAP, these data suggest that tumorigenic cancer cells are more susceptible to CAP treatment.
Figures

References
-
- Lord C. J. & Ashworth A. The DNA damage response and cancer therapy. Nature 481, 287–294 (2012). - PubMed
-
- Connell P. P. & Hellman S. Advances in Radiotherapy and Implications for the Next Century: A Historical Perspective. Cancer Res 69, 383–392 (2009). - PubMed
-
- Schwartz G. K. & Shah M. A. Targeting the Cell Cycle: A New Approach to Cancer Therapy. J Clin Oncol 23, 9408–9421 (2005). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

