Complete genome sequence of Saccharothrix espanaensis DSM 44229(T) and comparison to the other completely sequenced Pseudonocardiaceae
- PMID: 22958348
- PMCID: PMC3469384
- DOI: 10.1186/1471-2164-13-465
Complete genome sequence of Saccharothrix espanaensis DSM 44229(T) and comparison to the other completely sequenced Pseudonocardiaceae
Abstract
Background: The genus Saccharothrix is a representative of the family Pseudonocardiaceae, known to include producer strains of a wide variety of potent antibiotics. Saccharothrix espanaensis produces both saccharomicins A and B of the promising new class of heptadecaglycoside antibiotics, active against both bacteria and yeast.
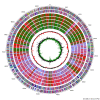
Results: To better assess its capabilities, the complete genome sequence of S. espanaensis was established. With a size of 9,360,653 bp, coding for 8,501 genes, it stands alongside other Pseudonocardiaceae with large genomes. Besides a predicted core genome of 810 genes shared in the family, S. espanaensis has a large number of accessory genes: 2,967 singletons when compared to the family, of which 1,292 have no clear orthologs in the RefSeq database. The genome analysis revealed the presence of 26 biosynthetic gene clusters potentially encoding secondary metabolites. Among them, the cluster coding for the saccharomicins could be identified.
Conclusion: S. espanaensis is the first completely sequenced species of the genus Saccharothrix. The genome discloses the cluster responsible for the biosynthesis of the saccharomicins, the largest oligosaccharide antibiotic currently identified. Moreover, the genome revealed 25 additional putative secondary metabolite gene clusters further suggesting the strain's potential for natural product synthesis.
Figures





References
-
- Labeda DP, Kroppenstedt RM. Phylogenetic analysis of Saccharothrix and related taxa: proposal for Actinosynnemataceae fam. nov. Int J Syst Bacteriol. 2000;50:331–336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

