How variable clones build an invariant retina
- PMID: 22958820
- PMCID: PMC3485567
- DOI: 10.1016/j.neuron.2012.06.033
How variable clones build an invariant retina
Abstract
A fundamental question in developmental neuroscience is how a collection of progenitor cells proliferates and differentiates to create a brain of the appropriate size and cellular composition. To address this issue, we devised lineage-tracing assays in developing zebrafish embryos to reconstruct entire retinal lineage progressions in vivo and thereby provide a complete quantitative map of the generation of a vertebrate CNS tissue from individual progenitors. These lineage data are consistent with a simple model in which the retina is derived from a set of equipotent retinal progenitor cells (RPCs) that are subject to stochastic factors controlling lineage progression. Clone formation in mutant embryos reveals that the transcription factor Ath5 acts as a molecular link between fate choice and mode of cell division, giving insight into the elusive molecular mechanisms of histogenesis, the conserved temporal order by which neurons of different types exit the cell cycle.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
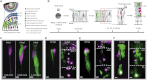
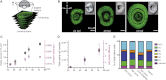

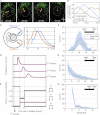


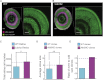
Comment in
-
Deterministic or stochastic choices in retinal neuron specification.Neuron. 2012 Sep 6;75(5):739-42. doi: 10.1016/j.neuron.2012.08.008. Neuron. 2012. PMID: 22958814 Free PMC article.
References
-
- Agathocleous M., Harris W.A. From progenitors to differentiated cells in the vertebrate retina. Annu. Rev. Cell Dev. Biol. 2009;25:45–69. - PubMed
-
- Alexiades M.R., Cepko C.L. Subsets of retinal progenitors display temporally regulated and distinct biases in the fates of their progeny. Development. 1997;124:1119–1131. - PubMed
-
- Angevine J.B., Jr., Sidman R.L. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. - PubMed
-
- Belecky-Adams T., Cook B., Adler R. Correlations between terminal mitosis and differentiated fate of retinal precursor cells in vivo and in vitro: analysis with the “window-labeling” technique. Dev. Biol. 1996;178:304–315. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

