A phylogenomic analysis of Escherichia coli / Shigella group: implications of genomic features associated with pathogenicity and ecological adaptation
- PMID: 22958895
- PMCID: PMC3444427
- DOI: 10.1186/1471-2148-12-174
A phylogenomic analysis of Escherichia coli / Shigella group: implications of genomic features associated with pathogenicity and ecological adaptation
Abstract
Background: The Escherichia coli species contains a variety of commensal and pathogenic strains, and its intraspecific diversity is extraordinarily high. With the availability of an increasing number of E. coli strain genomes, a more comprehensive concept of their evolutionary history and ecological adaptation can be developed using phylogenomic analyses. In this study, we constructed two types of whole-genome phylogenies based on 34 E. coli strains using collinear genomic segments. The first phylogeny was based on the concatenated collinear regions shared by all of the studied genomes, and the second phylogeny was based on the variable collinear regions that are absent from at least one genome. Intuitively, the first phylogeny is likely to reveal the lineal evolutionary history among these strains (i.e., an evolutionary phylogeny), whereas the latter phylogeny is likely to reflect the whole-genome similarities of extant strains (i.e., a similarity phylogeny).
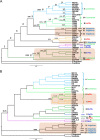
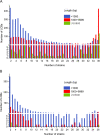
Results: Within the evolutionary phylogeny, the strains were clustered in accordance with known phylogenetic groups and phenotypes. When comparing evolutionary and similarity phylogenies, a concept emerges that Shigella may have originated from at least three distinct ancestors and evolved into a single clade. By scrutinizing the properties that are shared amongst Shigella strains but missing in other E. coli genomes, we found that the common regions of the Shigella genomes were mainly influenced by mobile genetic elements, implying that they may have experienced convergent evolution via horizontal gene transfer. Based on an inspection of certain key branches of interest, we identified several collinear regions that may be associated with the pathogenicity of specific strains. Moreover, by examining the annotated genes within these regions, further detailed evidence associated with pathogenicity was revealed.
Conclusions: Collinear regions are reliable genomic features used for phylogenomic analysis among closely related genomes while linking the genomic diversity with phenotypic differences in a meaningful way. The pathogenicity of a strain may be associated with both the arrival of virulence factors and the modification of genomes via mutations. Such phylogenomic studies that compare collinear regions of whole genomes will help to better understand the evolution and adaptation of closely related microbes and E. coli in particular.
Figures

References
-
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O. et al.Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5(1):e1000344. doi: 10.1371/journal.pgen.1000344. - DOI - PMC - PubMed
-
- Donnenberg MS. Escherichia coli: virulence mechanisms of a versatile pathogen. Academic Press: Elsevier Science; 2002.
-
- Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci USA. 2009;106(42):17939. doi: 10.1073/pnas.0903585106. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

