Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells
- PMID: 22959270
- PMCID: PMC3635072
- DOI: 10.1016/j.molcel.2012.08.002
Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells
Abstract
Triplex structure-forming GAA/TTC repeats pose a dual threat to the eukaryotic genome integrity. Their potential to expand can lead to gene inactivation, the cause of Friedreich's ataxia disease in humans. In model systems, long GAA/TTC tracts also act as chromosomal fragile sites that can trigger gross chromosomal rearrangements. The mechanisms that regulate the metabolism of GAA/TTC repeats are poorly understood. We have developed an experimental system in the yeast Saccharomyces cerevisiae that allows us to systematically identify genes crucial for maintaining the repeat stability. Two major groups of mutants defective in DNA replication or transcription initiation are found to be prone to fragility and large-scale expansions. We demonstrate that problems imposed by the repeats during DNA replication in actively dividing cells and during transcription initiation in nondividing cells can culminate in genome instability. We propose that similar mechanisms can mediate detrimental metabolism of GAA/TTC tracts in human cells.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
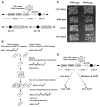


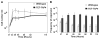

References
-
- Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. - PubMed
-
- Bellí G, Garí E, Aldea M, Herrero E. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast. 1998;14:1127–1138. - PubMed
-
- Campuzano V, Montermini L, Moltò MD, Pianese L, Cossée M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

