Ancient admixture in human history
- PMID: 22960212
- PMCID: PMC3522152
- DOI: 10.1534/genetics.112.145037
Ancient admixture in human history
Abstract
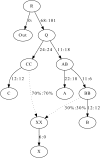
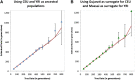
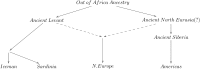
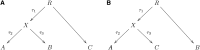
Population mixture is an important process in biology. We present a suite of methods for learning about population mixtures, implemented in a software package called ADMIXTOOLS, that support formal tests for whether mixture occurred and make it possible to infer proportions and dates of mixture. We also describe the development of a new single nucleotide polymorphism (SNP) array consisting of 629,433 sites with clearly documented ascertainment that was specifically designed for population genetic analyses and that we genotyped in 934 individuals from 53 diverse populations. To illustrate the methods, we give a number of examples that provide new insights about the history of human admixture. The most striking finding is a clear signal of admixture into northern Europe, with one ancestral population related to present-day Basques and Sardinians and the other related to present-day populations of northeast Asia and the Americas. This likely reflects a history of admixture between Neolithic migrants and the indigenous Mesolithic population of Europe, consistent with recent analyses of ancient bones from Sweden and the sequencing of the genome of the Tyrolean "Iceman."
Figures
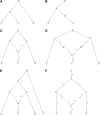
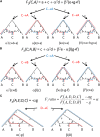
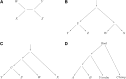
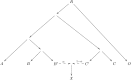


References
-
- Anthony D. W., 2007. The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World, Princeton University Press, Princeton, NJ.
-
- Barnard A., 1992. Hunters and Herders of Southern Africa. A comparative ethnography of the Khoisan peoples, Cambridge University Press, Cambridge, UK.
-
- Bramanti B., Thomas M. G., Haak W., Unterlaender M., Jores P., et al. 2009. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science 326: 137–140. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

