A novel approach for choosing summary statistics in approximate Bayesian computation
- PMID: 22960215
- PMCID: PMC3522150
- DOI: 10.1534/genetics.112.143164
A novel approach for choosing summary statistics in approximate Bayesian computation
Abstract
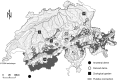
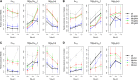
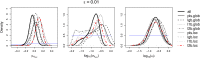
The choice of summary statistics is a crucial step in approximate Bayesian computation (ABC). Since statistics are often not sufficient, this choice involves a trade-off between loss of information and reduction of dimensionality. The latter may increase the efficiency of ABC. Here, we propose an approach for choosing summary statistics based on boosting, a technique from the machine-learning literature. We consider different types of boosting and compare them to partial least-squares regression as an alternative. To mitigate the lack of sufficiency, we also propose an approach for choosing summary statistics locally, in the putative neighborhood of the true parameter value. We study a demographic model motivated by the reintroduction of Alpine ibex (Capra ibex) into the Swiss Alps. The parameters of interest are the mean and standard deviation across microsatellites of the scaled ancestral mutation rate (θ(anc) = 4N(e)u) and the proportion of males obtaining access to matings per breeding season (ω). By simulation, we assess the properties of the posterior distribution obtained with the various methods. According to our criteria, ABC with summary statistics chosen locally via boosting with the L(2)-loss performs best. Applying that method to the ibex data, we estimate θ(anc)≈ 1.288 and find that most of the variation across loci of the ancestral mutation rate u is between 7.7 × 10(-4) and 3.5 × 10(-3) per locus per generation. The proportion of males with access to matings is estimated as ω≈ 0.21, which is in good agreement with recent independent estimates.
Figures



References
-
- Aeschbacher, A., 1978 Das Brunftverhalten des Alpensteinbocks. Eugen Rentsch Verlag, Erlenbach-Zürich, Switzerland.
-
- Akaike H., 1974. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19: 716–723.
-
- Beaumont M. A., 2010. Approximate Bayesian computation in evolution and ecology. Annu. Rev. Ecol. Evol. Syst. 41: 379–406.
-
- Beaumont M. A., Rannala B., 2004. The Bayesian revolution in genetics. Nat. Rev. Genet. 5: 251–261. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

