Alternative 3'-end processing of long noncoding RNA initiates construction of nuclear paraspeckles
- PMID: 22960638
- PMCID: PMC3474925
- DOI: 10.1038/emboj.2012.251
Alternative 3'-end processing of long noncoding RNA initiates construction of nuclear paraspeckles
Abstract
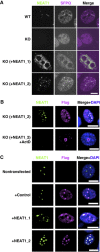
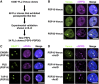
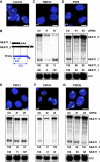
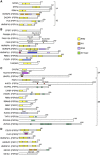
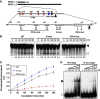
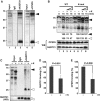
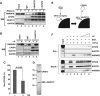
Paraspeckles are unique subnuclear structures built around a specific long noncoding RNA, NEAT1, which is comprised of two isoforms produced by alternative 3'-end processing (NEAT1_1 and NEAT1_2). To address the precise molecular processes that lead to paraspeckle formation, we identified 35 paraspeckle proteins (PSPs), mainly by colocalization screening with a fluorescent protein-tagged full-length cDNA library. Most of the newly identified PSPs possessed various putative RNA-binding domains. Subsequent RNAi analyses identified seven essential PSPs for paraspeckle formation. One of the essential PSPs, HNRNPK, appeared to affect the production of the essential NEAT1_2 isoform by negatively regulating the 3'-end polyadenylation of the NEAT1_1 isoform. An in vitro 3'-end processing assay revealed that HNRNPK arrested binding of the CPSF6-NUDT21 (CFIm) complex in the vicinity of the alternative polyadenylation site of NEAT1_1. In vitro binding assays showed that HNRNPK competed with CPSF6 for binding to NUDT21, which was the underlying mechanism to arrest CFIm binding by HNRNPK. This HNRNPK function led to the preferential accumulation of NEAT1_2 and initiated paraspeckle construction with multiple PSPs.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

