The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection
- PMID: 22960744
- PMCID: PMC3493121
- DOI: 10.1038/nature11353
The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection
Abstract
Several homology-dependent pathways can repair potentially lethal DNA double-strand breaks (DSBs). The first step common to all homologous recombination reactions is the 5'-3' degradation of DSB ends that yields the 3' single-stranded DNA required for the loading of checkpoint and recombination proteins. In yeast, the Mre11-Rad50-Xrs2 complex (Xrs2 is known as NBN or NBS1 in humans) and Sae2 (known as RBBP8 or CTIP in humans) initiate end resection, whereas long-range resection depends on the exonuclease Exo1, or the helicase-topoisomerase complex Sgs1-Top3-Rmi1 together with the endonuclease Dna2 (refs 1-6). DSBs occur in the context of chromatin, but how the resection machinery navigates through nucleosomal DNA is a process that is not well understood. Here we show that the yeast Saccharomyces cerevisiae Fun30 protein and its human counterpart SMARCAD1 (ref. 8), two poorly characterized ATP-dependent chromatin remodellers of the Snf2 ATPase family, are directly involved in the DSB response. Fun30 physically associates with DSB ends and directly promotes both Exo1- and Sgs1-dependent end resection through a mechanism involving its ATPase activity. The function of Fun30 in resection facilitates the repair of camptothecin-induced DNA lesions, although it becomes dispensable when Exo1 is ectopically overexpressed. Interestingly, SMARCAD1 is also recruited to DSBs, and the kinetics of recruitment is similar to that of EXO1. The loss of SMARCAD1 impairs end resection and recombinational DNA repair, and renders cells hypersensitive to DNA damage resulting from camptothecin or poly(ADP-ribose) polymerase inhibitor treatments. These findings unveil an evolutionarily conserved role for the Fun30 and SMARCAD1 chromatin remodellers in controlling end resection, homologous recombination and genome stability in the context of chromatin.
Figures
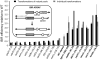

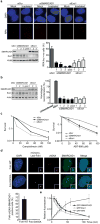
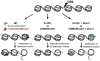
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

