IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress
- PMID: 22960911
- PMCID: PMC3480289
- DOI: 10.1105/tpc.112.097006
IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress
Abstract
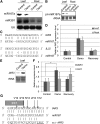
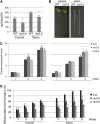
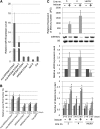
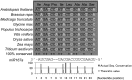
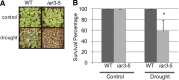
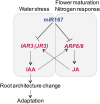
The functions of microRNAs and their target mRNAs in Arabidopsis thaliana development have been widely documented; however, roles of stress-responsive microRNAs and their targets are not as well understood. Using small RNA deep sequencing and ATH1 microarrays to profile mRNAs, we identified IAA-Ala Resistant3 (IAR3) as a new target of miR167a. As expected, IAR3 mRNA was cleaved at the miR167a complementary site and under high osmotic stress miR167a levels decreased, whereas IAR3 mRNA levels increased. IAR3 hydrolyzes an inactive form of auxin (indole-3-acetic acid [IAA]-alanine) and releases bioactive auxin (IAA), a central phytohormone for root development. In contrast with the wild type, iar3 mutants accumulated reduced IAA levels and did not display high osmotic stress-induced root architecture changes. Transgenic plants expressing a cleavage-resistant form of IAR3 mRNA accumulated high levels of IAR3 mRNAs and showed increased lateral root development compared with transgenic plants expressing wild-type IAR3. Expression of an inducible noncoding RNA to sequester miR167a by target mimicry led to an increase in IAR3 mRNA levels, further confirming the inverse relationship between the two partners. Sequence comparison revealed the miR167 target site on IAR3 mRNA is conserved in evolutionarily distant plant species. Finally, we showed that IAR3 is required for drought tolerance.
Figures
References
-
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Arenas-Huertero C., Pérez B., Rabanal F., Blanco-Melo D., De la Rosa C., Estrada-Navarrete G., Sanchez F., Covarrubias A.A., Reyes J.L. (2009). Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol. Biol. 70: 385–401 - PubMed
-
- Bartel D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

