Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize
- PMID: 22961134
- PMCID: PMC3490580
- DOI: 10.1104/pp.112.204743
Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize
Abstract
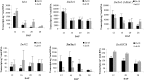
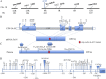
The phytohormone auxin (indole-3-acetic acid [IAA]) plays a fundamental role in vegetative and reproductive plant development. Here, we characterized a seed-specific viable maize (Zea mays) mutant, defective endosperm18 (de18) that is impaired in IAA biosynthesis. de18 endosperm showed large reductions of free IAA levels and is known to have approximately 40% less dry mass, compared with De18. Cellular analyses showed lower total cell number, smaller cell volume, and reduced level of endoreduplication in the mutant endosperm. Gene expression analyses of seed-specific tryptophan-dependent IAA pathway genes, maize Yucca1 (ZmYuc1), and two tryptophan-aminotransferase co-orthologs were performed to understand the molecular basis of the IAA deficiency in the mutant. Temporally, all three genes showed high expression coincident with high IAA levels; however, only ZmYuc1 correlated with the reduced IAA levels in the mutant throughout endosperm development. Furthermore, sequence analyses of ZmYuc1 complementary DNA and genomic clones revealed many changes specific to the mutant, including a 2-bp insertion that generated a premature stop codon and a truncated YUC1 protein of 212 amino acids, compared with the 400 amino acids in the De18. The putative, approximately 1.5-kb, Yuc1 promoter region also showed many rearrangements, including a 151-bp deletion in the mutant. Our concurrent high-density mapping and annotation studies of chromosome 10, contig 395, showed that the De18 locus was tightly linked to the gene ZmYuc1. Collectively, the data suggest that the molecular changes in the ZmYuc1 gene encoding the YUC1 protein are the causal basis of impairment in a critical step in IAA biosynthesis, essential for normal endosperm development in maize.
Figures



Similar articles
-
Sugar-hormone cross-talk in seed development: two redundant pathways of IAA biosynthesis are regulated differentially in the invertase-deficient miniature1 (mn1) seed mutant in maize.Mol Plant. 2010 Nov;3(6):1026-36. doi: 10.1093/mp/ssq057. Epub 2010 Oct 5. Mol Plant. 2010. PMID: 20924026
-
Transcriptomic and metabolomic analysis of ZmYUC1 mutant reveals the role of auxin during early endosperm formation in maize.Plant Sci. 2019 Apr;281:133-145. doi: 10.1016/j.plantsci.2019.01.027. Epub 2019 Feb 5. Plant Sci. 2019. PMID: 30824046
-
ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development.Plant Physiol. 2010 Mar;152(3):1373-90. doi: 10.1104/pp.109.150193. Epub 2009 Dec 31. Plant Physiol. 2010. PMID: 20044449 Free PMC article.
-
Auxin biosynthesis in maize.Plant Biol (Stuttg). 2006 May;8(3):334-9. doi: 10.1055/s-2006-923883. Plant Biol (Stuttg). 2006. PMID: 16807825 Review.
-
The pathway of auxin biosynthesis in plants.J Exp Bot. 2012 May;63(8):2853-72. doi: 10.1093/jxb/ers091. Epub 2012 Mar 23. J Exp Bot. 2012. PMID: 22447967 Review.
Cited by
-
The auxins, IAA and PAA, are synthesized by similar steps catalyzed by different enzymes.Plant Signal Behav. 2016 Nov;11(11):e1250993. doi: 10.1080/15592324.2016.1250993. Plant Signal Behav. 2016. PMID: 27808586 Free PMC article.
-
ENB1 encodes a cellulose synthase 5 that directs synthesis of cell wall ingrowths in maize basal endosperm transfer cells.Plant Cell. 2022 Mar 4;34(3):1054-1074. doi: 10.1093/plcell/koab312. Plant Cell. 2022. PMID: 34935984 Free PMC article.
-
Characterization of Imprinted Genes in Rice Reveals Conservation of Regulation and Imprinting with Other Plant Species.Plant Physiol. 2018 Aug;177(4):1754-1771. doi: 10.1104/pp.17.01621. Epub 2018 Jun 18. Plant Physiol. 2018. PMID: 29914891 Free PMC article.
-
Genome-wide identification and characterization of lncRNAs in sunflower endosperm.BMC Plant Biol. 2022 Oct 22;22(1):494. doi: 10.1186/s12870-022-03882-5. BMC Plant Biol. 2022. PMID: 36271333 Free PMC article.
-
Paternal imprinting of dosage-effect defective1 contributes to seed weight xenia in maize.Nat Commun. 2022 Sep 13;13(1):5366. doi: 10.1038/s41467-022-33055-9. Nat Commun. 2022. PMID: 36100609 Free PMC article.
References
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. (1993) Current Protocols in Molecular Biology. Wiley, New York
-
- Barlow PW. (1977) The time-course of endoreduplication of nuclear DNA in the root cap of Zea mays. Eur J Cell Biol 16: 98–105
-
- Bengough AG, Iijima M, Barlow PW. (2001) Image analysis of maize root caps: estimating cell numbers from 2-D longitudinal sections. Ann Bot (Lond) 87: 693–698
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

