Dxo1 is a new type of eukaryotic enzyme with both decapping and 5'-3' exoribonuclease activity
- PMID: 22961381
- PMCID: PMC3711404
- DOI: 10.1038/nsmb.2381
Dxo1 is a new type of eukaryotic enzyme with both decapping and 5'-3' exoribonuclease activity
Abstract
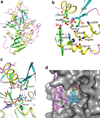
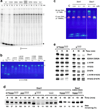
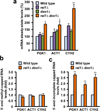
Recent studies showed that Rai1 is a crucial component of the mRNA 5'-end-capping quality-control mechanism in yeast. The yeast genome encodes a weak homolog of Rai1, Ydr370C, but little is known about this protein. Here we report the crystal structures of Ydr370C from Kluyveromyces lactis and the first biochemical and functional studies on this protein. The overall structure of Ydr370C is similar to Rai1. Ydr370C has robust decapping activity on RNAs with unmethylated caps, but it has no detectable pyrophosphohydrolase activity. Unexpectedly, Ydr370C also possesses distributive, 5'-3' exoRNase activity, and we propose the name Dxo1 for this new eukaryotic enzyme with both decapping and exonuclease activities. Studies of yeast in which both Dxo1 and Rai1 are disrupted reveal that mRNAs with incomplete caps are produced even under normal growth conditions, in sharp contrast to current understanding of the capping process.
Figures



References
-
- Shatkin AJ, Manley JL. The ends of the affair: capping and polyadenylation. Nature Struct. Biol. 2000;7:838–842. - PubMed
-
- Shuman S. The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny. Cold Spring Harbor Symp. Quant. Biol. LXVI. 2001:301–312. - PubMed
-
- Coller J, Parker R. Eukaryotic mRNA decapping. Ann. Rev. Biochem. 2004;73:861–890. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

