ALKBH1 is a histone H2A dioxygenase involved in neural differentiation
- PMID: 22961808
- PMCID: PMC3546389
- DOI: 10.1002/stem.1228
ALKBH1 is a histone H2A dioxygenase involved in neural differentiation
Abstract
AlkB homolog 1 (ALKBH1) is one of nine members of the family of mammalian AlkB homologs. Most Alkbh1(-/-) mice die during embryonic development, and survivors are characterized by defects in tissues originating from the ectodermal lineage. In this study, we show that deletion of Alkbh1 prolonged the expression of pluripotency markers in embryonic stem cells and delayed the induction of genes involved in early differentiation. In vitro differentiation to neural progenitor cells (NPCs) displayed an increased rate of apoptosis in the Alkbh1(-/-) NPCs when compared with wild-type cells. Whole-genome expression analysis and chromatin immunoprecipitation revealed that ALKBH1 regulates both directly and indirectly, a subset of genes required for neural development. Furthermore, our in vitro enzyme activity assays demonstrate that ALKBH1 is a histone dioxygenase that acts specifically on histone H2A. Mass spectrometric analysis demonstrated that histone H2A from Alkbh1(-/-) mice are improperly methylated. Our results suggest that ALKBH1 is involved in neural development by modifying the methylation status of histone H2A.
Copyright © 2012 AlphaMed Press.
Figures
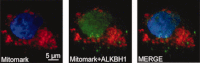
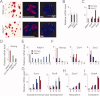




Similar articles
-
Role of ALKBH1 in the Core Transcriptional Network of Embryonic Stem Cells.Cell Physiol Biochem. 2016;38(1):173-84. doi: 10.1159/000438619. Epub 2016 Jan 15. Cell Physiol Biochem. 2016. PMID: 26765775
-
Characterization of human AlkB homolog 1 produced in mammalian cells and demonstration of mitochondrial dysfunction in ALKBH1-deficient cells.Biochem Biophys Res Commun. 2018 Jan 1;495(1):98-103. doi: 10.1016/j.bbrc.2017.10.158. Epub 2017 Oct 31. Biochem Biophys Res Commun. 2018. PMID: 29097205 Free PMC article.
-
Impaired placental trophoblast lineage differentiation in Alkbh1(-/-) mice.Dev Dyn. 2008 Feb;237(2):316-27. doi: 10.1002/dvdy.21418. Dev Dyn. 2008. PMID: 18163532
-
Demethyltransferase AlkBH1 substrate diversity and relationship to human diseases.Mol Biol Rep. 2021 May;48(5):4747-4756. doi: 10.1007/s11033-021-06421-x. Epub 2021 May 27. Mol Biol Rep. 2021. PMID: 34046849 Review.
-
Histone variants as emerging regulators of embryonic stem cell identity.Epigenetics. 2015;10(7):563-73. doi: 10.1080/15592294.2015.1053682. Epigenetics. 2015. PMID: 26114724 Free PMC article. Review.
Cited by
-
Differential repair of etheno-DNA adducts by bacterial and human AlkB proteins.DNA Repair (Amst). 2015 Jun;30:1-10. doi: 10.1016/j.dnarep.2015.02.021. Epub 2015 Mar 5. DNA Repair (Amst). 2015. PMID: 25797601 Free PMC article.
-
Dealing with an Unconventional Genetic Code in Mitochondria: The Biogenesis and Pathogenic Defects of the 5-Formylcytosine Modification in Mitochondrial tRNAMet.Biomolecules. 2017 Mar 2;7(1):24. doi: 10.3390/biom7010024. Biomolecules. 2017. PMID: 28257121 Free PMC article. Review.
-
ALKBH4-dependent demethylation of actin regulates actomyosin dynamics.Nat Commun. 2013;4:1832. doi: 10.1038/ncomms2863. Nat Commun. 2013. PMID: 23673617 Free PMC article.
-
Spectroscopic and in vitro Investigations of Fe2+ /α-Ketoglutarate-Dependent Enzymes Involved in Nucleic Acid Repair and Modification.Chembiochem. 2022 Jun 3;23(11):e202100605. doi: 10.1002/cbic.202100605. Epub 2022 Feb 15. Chembiochem. 2022. PMID: 35040547 Free PMC article. Review.
-
Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome.Nucleic Acids Res. 2018 Dec 14;46(22):11659-11670. doi: 10.1093/nar/gky1104. Nucleic Acids Res. 2018. PMID: 30412255 Free PMC article.
References
-
- Loebel DA, Watson CM, De Young RA, et al. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev Biol. 2003;264:1–14. - PubMed
-
- Smith AG. Embryo-derived stem cells: Of mice and men. Annu Rev Cell Dev Biol. 2001;17:435–462. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. - PubMed
-
- Masui S, Nakatake Y, Toyooka Y, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

