Pleiotropic control of secondary metabolism and morphological development by KsbC, a butyrolactone autoregulator receptor homologue in Kitasatospora setae
- PMID: 22961899
- PMCID: PMC3485943
- DOI: 10.1128/AEM.02355-12
Pleiotropic control of secondary metabolism and morphological development by KsbC, a butyrolactone autoregulator receptor homologue in Kitasatospora setae
Abstract
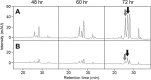
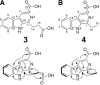
The γ-butyrolactone autoregulator signaling cascades have been shown to control secondary metabolism and/or morphological development among many Streptomyces species. However, the conservation and variation of the regulatory systems among actinomycetes remain to be clarified. The genome sequence of Kitasatospora setae, which also belongs to the family Streptomycetaceae containing the genus Streptomyces, has revealed the presence of three homologues of the autoregulator receptor: KsbA, which has previously been confirmed to be involved only in secondary metabolism; KsbB; and KsbC. We describe here the characterization of ksbC, whose regulatory cluster closely resembles the Streptomyces virginiae barA locus responsible for the autoregulator signaling cascade. Deletion of the gene ksbC resulted in lowered production of bafilomycin and a defect of aerial mycelium formation, together with the early and enhanced production of a novel β-carboline alkaloid named kitasetaline. A putative kitasetaline biosynthetic gene cluster was identified, and its expression in a heterologous host led to the production of kitasetaline together with JBIR-133, the production of which is also detected in the ksbC disruptant, and JBIR-134 as novel β-carboline alkaloids, indicating that these genes were biosynthetic genes for β-carboline alkaloid and thus are the first such genes to be discovered in bacteria.
Figures




References
-
- Aroonsri A, Kitani S, Ikeda H, Nihira T. 2012. Kitasetaline, a novel β-carboline alkaloid from Kitasatospora setae NBRC 14216T. J. Biosci. Bioeng. 114:56–58 - PubMed
-
- Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208–215 - PubMed
-
- Bierman M, et al. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

