Pruning rogue taxa improves phylogenetic accuracy: an efficient algorithm and webservice
- PMID: 22962004
- PMCID: PMC3526802
- DOI: 10.1093/sysbio/sys078
Pruning rogue taxa improves phylogenetic accuracy: an efficient algorithm and webservice
Abstract
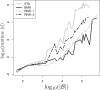
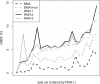
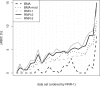
The presence of rogue taxa (rogues) in a set of trees can frequently have a negative impact on the results of a bootstrap analysis (e.g., the overall support in consensus trees). We introduce an efficient graph-based algorithm for rogue taxon identification as well as an interactive webservice implementing this algorithm. Compared with our previous method, the new algorithm is up to 4 orders of magnitude faster, while returning qualitatively identical results. Because of this significant improvement in scalability, the new algorithm can now identify substantially more complex and compute-intensive rogue taxon constellations. On a large and diverse collection of real-world data sets, we show that our method yields better supported reduced/pruned consensus trees than any competing rogue taxon identification method. Using the parallel version of our open-source code, we successfully identified rogue taxa in a set of 100 trees with 116 334 taxa each. For simulated data sets, we show that when removing/pruning rogue taxa with our method from a tree set, we consistently obtain bootstrap consensus trees as well as maximum-likelihood trees that are topologically closer to the respective true trees.
Figures
References
-
- Aberer A.J., Stamatakis A. A simple and accurate method for rogue taxon identification. IEEE International Conference on Bioinformatics and Biomedicine; Atlanta (GA). 2011. pp. 118–122. IEEE.
-
- Bryant D. A classification of consensus methods for phylogenetics. DIMACS Series in Discrete Mathematics and Theoretical Computer Science. 2003;61:163–184.
-
- Dunn C.W., Hejnol A., Matus D.Q., Pang K., Browne W.E., Smith S.A., Seaver E., Rouse G.W., Obst M., Edgecombe G.D., Sørensen M.V., Haddock S.H.D., Schmidt-Rhaesa A., Okusu A., Kristensen R.M., Wheeler W.C., Martindale M.Q., Giribet G. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. - PubMed
-
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. - PubMed
-
- Maddison W., Maddison D. Mesquite: a modular system for evolutionary analysis. Evolution. 2008;62:1103–1118. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

