Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer
- PMID: 22962056
- PMCID: PMC3518128
- DOI: 10.1074/mcp.O112.019802
Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer
Abstract
There is an immediate need for improved methods to systematically and precisely quantify large sets of peptides in complex biological samples. To date protein quantification in biological samples has been routinely performed on triple quadrupole instruments operated in selected reaction monitoring mode (SRM), and two major challenges remain. Firstly, the number of peptides to be included in one survey experiment needs to be increased to routinely reach several hundreds, and secondly, the degree of selectivity should be improved so as to reliably discriminate the targeted analytes from background interferences. High resolution and accurate mass (HR/AM) analysis on the recently developed Q-Exactive mass spectrometer can potentially address these issues. This instrument presents a unique configuration: it is constituted of an orbitrap mass analyzer equipped with a quadrupole mass filter as the front-end for precursor ion mass selection. This configuration enables new quantitative methods based on HR/AM measurements, including targeted analysis in MS mode (single ion monitoring) and in MS/MS mode (parallel reaction monitoring). The ability of the quadrupole to select a restricted m/z range allows one to overcome the dynamic range limitations associated with trapping devices, and the MS/MS mode provides an additional stage of selectivity. When applied to targeted protein quantification in urine samples and benchmarked with the reference SRM technique, the quadrupole-orbitrap instrument exhibits similar or better performance in terms of selectivity, dynamic range, and sensitivity. This high performance is further enhanced by leveraging the multiplexing capability of the instrument to design novel acquisition methods and apply them to large targeted proteomic studies for the first time, as demonstrated on 770 tryptic yeast peptides analyzed in one 60-min experiment. The increased quality of quadrupole-orbitrap data has the potential to improve existing protein quantification methods in complex samples and address the pressing demand of systems biology or biomarker evaluation studies.
Figures
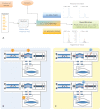


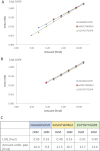
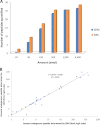
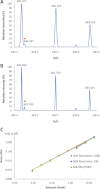
Similar articles
-
Technical considerations for large-scale parallel reaction monitoring analysis.J Proteomics. 2014 Apr 4;100:147-59. doi: 10.1016/j.jprot.2013.10.029. Epub 2013 Nov 4. J Proteomics. 2014. PMID: 24200835
-
Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer.Mol Cell Proteomics. 2011 Sep;10(9):M111.011015. doi: 10.1074/mcp.M111.011015. Epub 2011 Jun 3. Mol Cell Proteomics. 2011. PMID: 21642640 Free PMC article.
-
Parallel reaction monitoring using quadrupole-Orbitrap mass spectrometer: Principle and applications.Proteomics. 2016 Aug;16(15-16):2146-59. doi: 10.1002/pmic.201500543. Epub 2016 May 27. Proteomics. 2016. PMID: 27145088 Review.
-
A review on mass spectrometry-based quantitative proteomics: Targeted and data independent acquisition.Anal Chim Acta. 2017 Apr 29;964:7-23. doi: 10.1016/j.aca.2017.01.059. Epub 2017 Feb 2. Anal Chim Acta. 2017. PMID: 28351641 Review.
-
Large-Scale Targeted Proteomics Using Internal Standard Triggered-Parallel Reaction Monitoring (IS-PRM).Mol Cell Proteomics. 2015 Jun;14(6):1630-44. doi: 10.1074/mcp.O114.043968. Epub 2015 Mar 9. Mol Cell Proteomics. 2015. PMID: 25755295 Free PMC article.
Cited by
-
Oncogenic KRAS and BRAF Drive Metabolic Reprogramming in Colorectal Cancer.Mol Cell Proteomics. 2016 Sep;15(9):2924-38. doi: 10.1074/mcp.M116.058925. Epub 2016 Jun 23. Mol Cell Proteomics. 2016. PMID: 27340238 Free PMC article.
-
Determination of Site-Specific Phosphorylation Occupancy Using Targeted Mass Spectrometry Reveals the Regulation of Human Apical Bile Acid Transporter, ASBT.ACS Omega. 2024 Aug 30;9(37):38477-38489. doi: 10.1021/acsomega.4c02999. eCollection 2024 Sep 17. ACS Omega. 2024. PMID: 39310206 Free PMC article.
-
Label-Free Proteomic Analysis of Molecular Effects of 2-Methoxy-1,4-naphthoquinone on Penicillium italicum.Int J Mol Sci. 2019 Jul 14;20(14):3459. doi: 10.3390/ijms20143459. Int J Mol Sci. 2019. PMID: 31337149 Free PMC article.
-
Identification of Unique Peptides for SARS-CoV-2 Diagnostics and Vaccine Development by an In Silico Proteomics Approach.Front Immunol. 2021 Sep 24;12:725240. doi: 10.3389/fimmu.2021.725240. eCollection 2021. Front Immunol. 2021. PMID: 34630400 Free PMC article.
-
Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses.Chem Rev. 2018 Sep 12;118(17):7886-7930. doi: 10.1021/acs.chemrev.7b00732. Epub 2018 Mar 19. Chem Rev. 2018. PMID: 29553244 Free PMC article. Review.
References
-
- Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 - PubMed
-
- Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 - PubMed
-
- Aebersold R. (2009) A stress test for mass spectrometry-based proteomics. Nat. Methods 6, 411–412 - PubMed
-
- Domon B., Aebersold R. (2010) Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 28, 710–721 - PubMed
-
- Anderson N. L., Anderson N. G. (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

