Understanding cullin-RING E3 biology through proteomics-based substrate identification
- PMID: 22962057
- PMCID: PMC3518111
- DOI: 10.1074/mcp.R112.021154
Understanding cullin-RING E3 biology through proteomics-based substrate identification
Abstract
Protein turnover through the ubiquitin-proteasome pathway controls numerous developmental decisions and biochemical processes in eukaryotes. Central to protein ubiquitylation are ubiquitin ligases, which provide specificity in targeted ubiquitylation. With more than 600 ubiquitin ligases encoded by the human genome, many of which remain to be studied, considerable effort is being placed on the development of methods for identifying substrates of specific ubiquitin ligases. In this review, we describe proteomic technologies for the identification of ubiquitin ligase targets, with a particular focus on members of the cullin-RING E3 class of ubiquitin ligases, which use F-box proteins as substrate specific adaptor proteins. Various proteomic methods are described and are compared with genetic approaches that are available. The continued development of such methods is likely to have a substantial impact on the ubiquitin-proteasome field.
Conflict of interest statement
Conflict of interest statement: J.W.H. is a consultant for Millennium Pharmaceuticals.
Figures

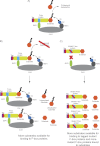
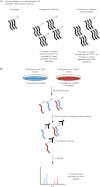
Similar articles
-
Cullin-RING E3 Ubiquitin Ligases: Bridges to Destruction.Subcell Biochem. 2017;83:323-347. doi: 10.1007/978-3-319-46503-6_12. Subcell Biochem. 2017. PMID: 28271482 Free PMC article. Review.
-
Cullin 5-RING E3 ubiquitin ligases, new therapeutic targets?Biochimie. 2016 Mar;122:339-47. doi: 10.1016/j.biochi.2015.08.003. Epub 2015 Aug 4. Biochimie. 2016. PMID: 26253693 Review.
-
Functional characterization of Cullin-1-RING ubiquitin ligase (CRL1) complex in Leishmania infantum.PLoS Pathog. 2024 Jul 17;20(7):e1012336. doi: 10.1371/journal.ppat.1012336. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 39018347 Free PMC article.
-
Coordinated Actions Between p97 and Cullin-RING Ubiquitin Ligases for Protein Degradation.Adv Exp Med Biol. 2020;1217:61-78. doi: 10.1007/978-981-15-1025-0_5. Adv Exp Med Biol. 2020. PMID: 31898222 Review.
-
Cullin-RING Ubiquitin Ligase Regulatory Circuits: A Quarter Century Beyond the F-Box Hypothesis.Annu Rev Biochem. 2021 Jun 20;90:403-429. doi: 10.1146/annurev-biochem-090120-013613. Epub 2021 Apr 6. Annu Rev Biochem. 2021. PMID: 33823649 Free PMC article. Review.
Cited by
-
The NEDD8 inhibitor MLN4924 increases the size of the nucleolus and activates p53 through the ribosomal-Mdm2 pathway.Oncogene. 2016 Jan 28;35(4):415-26. doi: 10.1038/onc.2015.104. Epub 2015 Apr 13. Oncogene. 2016. PMID: 25867069
-
Genome-Wide Analysis of U-box E3 Ubiquitin Ligase Family in Response to ABA Treatment in Salvia miltiorrhiza.Front Plant Sci. 2022 Feb 9;13:829447. doi: 10.3389/fpls.2022.829447. eCollection 2022. Front Plant Sci. 2022. PMID: 35222487 Free PMC article.
-
An E2-ubiquitin thioester-driven approach to identify substrates modified with ubiquitin and ubiquitin-like molecules.Nat Commun. 2018 Nov 14;9(1):4776. doi: 10.1038/s41467-018-07251-5. Nat Commun. 2018. PMID: 30429481 Free PMC article.
-
Protein Engineering in the Ubiquitin System: Tools for Discovery and Beyond.Pharmacol Rev. 2020 Apr;72(2):380-413. doi: 10.1124/pr.118.015651. Pharmacol Rev. 2020. PMID: 32107274 Free PMC article. Review.
-
Chromatin-Bound Cullin-Ring Ligases: Regulatory Roles in DNA Replication and Potential Targeting for Cancer Therapy.Front Mol Biosci. 2018 Mar 13;5:19. doi: 10.3389/fmolb.2018.00019. eCollection 2018. Front Mol Biosci. 2018. PMID: 29594129 Free PMC article. Review.
References
-
- Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 - PubMed
-
- Shabek N., Ciechanover A. (2010) Degradation of ubiquitin: the fate of the cellular reaper. Cell Cycle 9, 523–530 - PubMed
-
- Welcker M., Clurman B. E. (2008) FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

