Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation
- PMID: 22962365
- PMCID: PMC3488261
- DOI: 10.1093/nar/gks845
Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation
Abstract
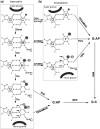
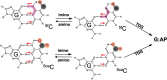
The mammalian thymine DNA glycosylase (TDG) is implicated in active DNA demethylation via the base excision repair pathway. TDG excises the mismatched base from G:X mismatches, where X is uracil, thymine or 5-hydroxymethyluracil (5hmU). These are, respectively, the deamination products of cytosine, 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC). In addition, TDG excises the Tet protein products 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) but not 5hmC and 5mC, when paired with a guanine. Here we present a post-reactive complex structure of the human TDG domain with a 28-base pair DNA containing a G:5hmU mismatch. TDG flips the target nucleotide from the double-stranded DNA, cleaves the N-glycosidic bond and leaves the C1' hydrolyzed abasic sugar in the flipped state. The cleaved 5hmU base remains in a binding pocket of the enzyme. TDG allows hydrogen-bonding interactions to both T/U-based (5hmU) and C-based (5caC) modifications, thus enabling its activity on a wider range of substrates. We further show that the TDG catalytic domain has higher activity for 5caC at a lower pH (5.5) as compared to the activities at higher pH (7.5 and 8.0) and that the structurally related Escherichia coli mismatch uracil glycosylase can excise 5caC as well. We discuss several possible mechanisms, including the amino-imino tautomerization of the substrate base that may explain how TDG discriminates against 5hmC and 5mC.
Figures





References
-
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

