Genomic context analysis reveals dense interaction network between vertebrate ultraconserved non-coding elements
- PMID: 22962458
- PMCID: PMC3436827
- DOI: 10.1093/bioinformatics/bts400
Genomic context analysis reveals dense interaction network between vertebrate ultraconserved non-coding elements
Abstract
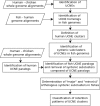
Motivation: Genomic context analysis, also known as phylogenetic profiling, is widely used to infer functional interactions between proteins but rarely applied to non-coding cis-regulatory DNA elements. We were wondering whether this approach could provide insights about utlraconserved non-coding elements (UCNEs). These elements are organized as large clusters, so-called gene regulatory blocks (GRBs) around key developmental genes. Their molecular functions and the reasons for their high degree of conservation remain enigmatic.
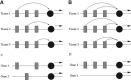
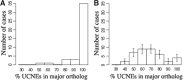
Results: In a special setting of genomic context analysis, we analyzed the fate of GRBs after a whole-genome duplication event in five fish genomes. We found that in most cases all UCNEs were retained together as a single block, whereas the corresponding target genes were often retained in two copies, one completely devoid of UCNEs. This 'winner-takes-all' pattern suggests that UCNEs of a GRB function in a highly cooperative manner. We propose that the multitude of interactions between UCNEs is the reason for their extreme sequence conservation.
Supplementary information: Supplementary data are available at Bioinformatics online and at http://ccg.vital-it.ch/ucne/
Figures
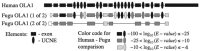
Similar articles
-
UCNEbase--a database of ultraconserved non-coding elements and genomic regulatory blocks.Nucleic Acids Res. 2013 Jan;41(Database issue):D101-9. doi: 10.1093/nar/gks1092. Epub 2012 Nov 27. Nucleic Acids Res. 2013. PMID: 23193254 Free PMC article.
-
Comparative genomics of ParaHox clusters of teleost fishes: gene cluster breakup and the retention of gene sets following whole genome duplications.BMC Genomics. 2007 Sep 6;8:312. doi: 10.1186/1471-2164-8-312. BMC Genomics. 2007. PMID: 17822543 Free PMC article.
-
Evolutionary conservation of DNA methylation in CpG sites within ultraconserved noncoding elements.Epigenetics. 2018;13(1):49-60. doi: 10.1080/15592294.2017.1411447. Epub 2018 Feb 6. Epigenetics. 2018. PMID: 29372669 Free PMC article.
-
Comparative genomics using fugu: a tool for the identification of conserved vertebrate cis-regulatory elements.Bioessays. 2005 Jan;27(1):100-7. doi: 10.1002/bies.20134. Bioessays. 2005. PMID: 15612032 Review.
-
Organization of conserved elements near key developmental regulators in vertebrate genomes.Adv Genet. 2008;61:307-38. doi: 10.1016/S0065-2660(07)00012-0. Adv Genet. 2008. PMID: 18282512 Review.
Cited by
-
Functional and structural basis of extreme conservation in vertebrate 5' untranslated regions.Nat Genet. 2021 May;53(5):729-741. doi: 10.1038/s41588-021-00830-1. Epub 2021 Apr 5. Nat Genet. 2021. PMID: 33821006 Free PMC article.
-
Overview of methods for characterization and visualization of a protein-protein interaction network in a multi-omics integration context.Front Mol Biosci. 2022 Sep 8;9:962799. doi: 10.3389/fmolb.2022.962799. eCollection 2022. Front Mol Biosci. 2022. PMID: 36158572 Free PMC article. Review.
-
Evolutionary stability of topologically associating domains is associated with conserved gene regulation.BMC Biol. 2018 Aug 7;16(1):87. doi: 10.1186/s12915-018-0556-x. BMC Biol. 2018. PMID: 30086749 Free PMC article.
-
Allele frequencies of variants in ultra conserved elements identify selective pressure on transcription factor binding.PLoS One. 2014 Nov 4;9(11):e110692. doi: 10.1371/journal.pone.0110692. eCollection 2014. PLoS One. 2014. PMID: 25369454 Free PMC article.
-
Genomic Characterization and Curation of UCEs Improves Species Tree Reconstruction.Syst Biol. 2021 Feb 10;70(2):307-321. doi: 10.1093/sysbio/syaa063. Syst Biol. 2021. PMID: 32750133 Free PMC article.
References
-
- Bejerano G., et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. - PubMed
-
- Calin G. A., et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. - PubMed
-
- Elgar G., Vavouri T. Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet. 2008;24:344–352. - PubMed

