Poly I:C enhances susceptibility to secondary pulmonary infections by gram-positive bacteria
- PMID: 22962579
- PMCID: PMC3433467
- DOI: 10.1371/journal.pone.0041879
Poly I:C enhances susceptibility to secondary pulmonary infections by gram-positive bacteria
Abstract
Secondary bacterial pneumonias are a frequent complication of influenza and other respiratory viral infections, but the mechanisms underlying viral-induced susceptibility to bacterial infections are poorly understood. In particular, it is unclear whether the host's response against the viral infection, independent of the injury caused by the virus, results in impairment of antibacterial host defense. Here, we sought to determine whether the induction of an "antiviral" immune state using various viral recognition receptor ligands was sufficient to result in decreased ability to combat common bacterial pathogens of the lung. Using a mouse model, animals were administered polyinosine-polycytidylic acid (poly I:C) or Toll-like 7 ligand (imiquimod or gardiquimod) intranasally, followed by intratracheal challenge with Streptococcus pneumoniae. We found that animals pre-exposed to poly I:C displayed impaired bacterial clearance and increased mortality. Poly I:C-exposed animals also had decreased ability to clear methicillin-resistant Staphylococcus aureus. Furthermore, we showed that activation of Toll-like receptor (TLR)3 and Retinoic acid inducible gene (RIG-I)/Cardif pathways, which recognize viral nucleic acids in the form of dsRNA, both contribute to poly I:C mediated impairment of bacterial clearance. Finally, we determined that poly I:C administration resulted in significant induction of type I interferons (IFNs), whereas the elimination of type I IFN signaling improved clearance and survival following secondary bacterial pneumonia. Collectively, these results indicate that in the lung, poly I:C administration is sufficient to impair pulmonary host defense against clinically important gram-positive bacterial pathogens, which appears to be mediated by type I IFNs.
Conflict of interest statement
Figures

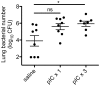
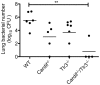
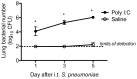
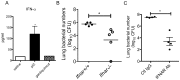

References
-
- Oliveira EC, Marik PE, Colice G (2001) Influenza pneumonia: a descriptive study. Chest 119: 1717–1723. - PubMed
-
- Korppi M (2002) Mixed microbial aetiology of community-acquired pneumonia in children. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica 110: 515–522. - PubMed
-
- Michelow IC, Olsen K, Lozano J, Rollins NK, Duffy LB, et al. (2004) Epidemiology and clinical characteristics of community-acquired pneumonia in hospitalized children. Pediatrics 113: 701–707. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

