The NAD(+)-dependent protein deacetylase activity of SIRT1 is regulated by its oligomeric status
- PMID: 22962634
- PMCID: PMC3435561
- DOI: 10.1038/srep00640
The NAD(+)-dependent protein deacetylase activity of SIRT1 is regulated by its oligomeric status
Abstract
SIRT1, a NAD(+)-dependent protein deacetylase, is an important regulator in cellular stress response and energy metabolism. While the list of SIRT1 substrates is growing, how the activity of SIRT1 is regulated remains unclear. We have previously reported that SIRT1 is activated by phosphorylation at a conserved Thr522 residue in response to environmental stress. Here we demonstrate that phosphorylation of Thr522 activates SIRT1 through modulation of its oligomeric status. We provide evidence that nonphosphorylated SIRT1 protein is aggregation-prone in vitro and in cultured cells. Conversely, phosphorylated SIRT1 protein is largely in the monomeric state and more active. Our findings reveal a novel mechanism for environmental regulation of SIRT1 activity, which may have important implications in understanding the molecular mechanism of stress response, cell survival, and aging.
Figures
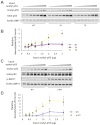

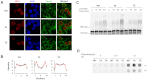
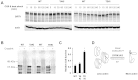
References
-
- Blander G. & Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem 73, 417–35 (2004). - PubMed
-
- Frye R. A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273, 793–8 (2000). - PubMed
-
- Kwon H. S. & Ott M. The ups and downs of SIRT1. Trends Biochem Sci 33, 517–25 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

