Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana
- PMID: 22962679
- PMCID: PMC3467300
- DOI: 10.1093/jxb/ers239
Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana
Abstract
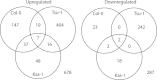
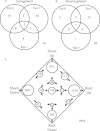
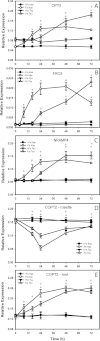
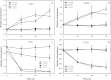
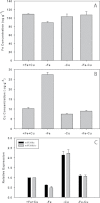
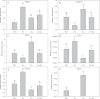
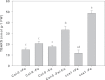
Iron (Fe) is an essential plant micronutrient, and its deficiency limits plant growth and development on alkaline soils. Under Fe deficiency, plant responses include up-regulation of genes involved in Fe uptake from the soil. However, little is known about shoot responses to Fe deficiency. Using microarrays to probe gene expression in Kas-1 and Tsu-1 ecotypes of Arabidopsis thaliana, and comparison with existing Col-0 data, revealed conserved rosette gene expression responses to Fe deficiency. Fe-regulated genes included known metal homeostasis-related genes, and a number of genes of unknown function. Several genes responded to Fe deficiency in both roots and rosettes. Fe deficiency led to up-regulation of Cu,Zn superoxide dismutase (SOD) genes CSD1 and CSD2, and down-regulation of FeSOD genes FSD1 and FSD2. Eight microRNAs were found to respond to Fe deficiency. Three of these (miR397a, miR398a, and miR398b/c) are known to regulate transcripts of Cu-containing proteins, and were down-regulated by Fe deficiency, suggesting that they could be involved in plant adaptation to Fe limitation. Indeed, Fe deficiency led to accumulation of Cu in rosettes, prior to any detectable decrease in Fe concentration. ccs1 mutants that lack functional Cu,ZnSOD proteins were prone to greater oxidative stress under Fe deficiency, indicating that increased Cu concentration under Fe limitation has an important role in oxidative stress prevention. The present results show that Cu accumulation, microRNA regulation, and associated differential expression of Fe and CuSOD genes are coordinated responses to Fe limitation.
Figures


References
-
- Beauclair L, Yu A, Bouche N. 2010. microRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis The Plant Journal 62 454––462 - PubMed
-
- Bonnet E, He Y, Billiau K, Van de Peer Y. 2010. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics Bioinformatics 26 1566––1568 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

