Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors
- PMID: 22963237
- PMCID: PMC3468123
- DOI: 10.1021/ja3065667
Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors
Abstract
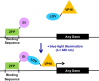
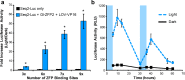
Advanced gene regulatory systems are necessary for scientific research, synthetic biology, and gene-based medicine. An ideal system would allow facile spatiotemporal manipulation of gene expression within a cell population that is tunable, reversible, repeatable, and can be targeted to diverse DNA sequences. To meet these criteria, a gene regulation system was engineered that combines light-sensitive proteins and programmable zinc finger transcription factors. This system, light-inducible transcription using engineered zinc finger proteins (LITEZ), uses two light-inducible dimerizing proteins from Arabidopsis thaliana, GIGANTEA and the LOV domain of FKF1, to control synthetic zinc finger transcription factor activity in human cells. Activation of gene expression in human cells engineered with LITEZ was reversible and repeatable by modulating the duration of illumination. The level of gene expression could also be controlled by modulating light intensity. Finally, gene expression could be activated in a spatially defined pattern by illuminating the human cell culture through a photomask of arbitrary geometry. LITEZ enables new approaches for precisely regulating gene expression in biotechnology and medicine, as well as studying gene function, cell-cell interactions, and tissue morphogenesis.
Figures


References
-
- Tigges M.; Fussenegger M. Curr. Opin. Biotechnol. 2009, 20, 449. - PubMed
-
- Gossen M.; Freundlieb S.; Bender G.; Muller G.; Hillen W.; Bujard H. Science 1995, 268, 1766. - PubMed
-
- No D.; Yao T. P.; Evans R. M. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 3346. - PMC - PubMed
- Weber W.; et al. Nat. Biotechnol. 2002, 20, 901. - PubMed
- Fussenegger M.; Morris R. P.; Fux C.; Rimann M.; von Stockar B.; Thompson C. J.; Bailey J. E. Nat. Biotechnol. 2000, 18, 1203. - PubMed
- Gitzinger M.; Kemmer C.; El-Baba M. D.; Weber W.; Fussenegger M. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 10638. - PMC - PubMed
- Rivera V. M.; et al. Nat. Med. 1996, 2, 1028. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

