Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts
- PMID: 22964225
- PMCID: PMC3518138
- DOI: 10.1074/mcp.M112.019984
Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts
Abstract
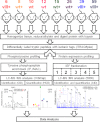
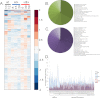
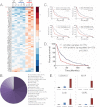
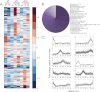
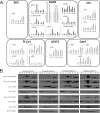
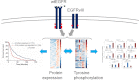
Glioblastoma multiforme (GBM) is a malignant primary brain tumor with a mean survival of 15 months with the current standard of care. Genetic profiling efforts have identified the amplification, overexpression, and mutation of the wild-type (wt) epidermal growth factor receptor tyrosine kinase (EGFR) in ≈ 50% of GBM patients. The genetic aberration of wtEGFR is frequently accompanied by the overexpression of a mutant EGFR known as EGFR variant III (EGFRvIII, de2-7EGFR, ΔEGFR), which is expressed in 30% of GBM tumors. The molecular mechanisms of tumorigenesis driven by EGFRvIII overexpression in human tumors have not been fully elucidated. To identify specific therapeutic targets for EGFRvIII driven tumors, it is important to gather a broad understanding of EGFRvIII specific signaling. Here, we have characterized signaling through the quantitative analysis of protein expression and tyrosine phosphorylation across a panel of glioblastoma tumor xenografts established from patient surgical specimens expressing wtEGFR or overexpressing wtEGFR (wtEGFR+) or EGFRvIII (EGFRvIII+). S100A10 (p11), major vault protein, guanylate-binding protein 1(GBP1), and carbonic anhydrase III (CAIII) were identified to have significantly increased expression in EGFRvIII expressing xenograft tumors relative to wtEGFR xenograft tumors. Increased expression of these four individual proteins was found to be correlated with poor survival in patients with GBM; the combination of these four proteins represents a prognostic signature for poor survival in gliomas. Integration of protein expression and phosphorylation data has uncovered significant heterogeneity among the various tumors and has highlighted several novel pathways, related to EGFR trafficking, activated in glioblastoma. The pathways and proteins identified in these tumor xenografts represent potential therapeutic targets for this disease.
Figures



References
-
- Furnari F. B., Fenton T., Bachoo R. M., Mukasa A., Stommel J. M., Stegh A., Hahn W. C., Ligon K. L., Louis D. N., Brennan C., Chin L., DePinho R. A., Cavenee W. K. (2007) Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 21, 2683–2710 - PubMed
-
- Stupp R., Mason W. P., van den Bent M. J., Weller M., Fisher B., Taphoorn M. J. B., Belanger K., Brandes A. A., Marosi C., Bogdahn U., Curschmann J., Janzer R. C., Ludwin S. K., Gorlia T., Allgeier A., Lacombe D., Cairncross J. G., Eisenhauer E., Mirimanoff R. O. (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 - PubMed
-
- Kagawa N., Maruno M., Suzuki T., Hashiba T., Hashimoto N., Izumoto S., Yoshimine T. (2006) Detection of genetic and chromosomal aberrations in medulloblastomas and primitive neuroectodermal tumors with DNA microarrays. Brain Tumor Pathol. 23, 41–47 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

