RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination
- PMID: 22964643
- PMCID: PMC5730454
- DOI: 10.1038/onc.2012.391
RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination
Abstract
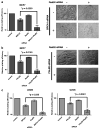
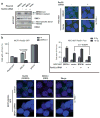
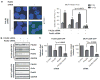
Synthetic lethality is an approach to study selective cell killing based on genotype. Previous work in our laboratory has shown that loss of RAD52 is synthetically lethal with BRCA2 deficiency, while exhibiting no impact on cell growth and viability in BRCA2-proficient cells. We now show that this same synthetically lethal relationship is evident in cells with deficiencies in BRCA1 or PALB2, which implicates BRCA1, PALB2 and BRCA2 in an epistatic relationship with one another. When RAD52 was depleted in BRCA1- or PALB2-deficient cells, a severe reduction in plating efficiency was observed, with many abortive attempts at cell division apparent in the double-depleted background. In contrast, when RAD52 was depleted in a BRCA1- or PALB2-wildtype background, a negligible decrease in colony survival was observed. The frequency of ionizing radiation-induced RAD51 foci formation and double-strand break-induced homologous recombination (HR) was decreased by 3- and 10-fold, respectively, when RAD52 was knocked down in BRCA1- or PALB2-depleted cells, with minimal effect in BRCA1- or PALB2-proficient cells. RAD52 function was independent of BRCA1 status, as evidenced by the lack of any defect in RAD52 foci formation in BRCA1-depleted cells. Collectively, these findings suggest that RAD52 is an alternative repair pathway of RAD51-mediated HR, and a target for therapy in cells deficient in the BRCA1-PALB2-BRCA2 repair pathway.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Iglehart JD, Silver DP. Synthetic Lethality — A New Direction in Cancer-Drug Development. N Engl J Med. 2009;361:189–191. - PubMed
-
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. - PubMed
-
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. - PubMed
-
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

