The selfish Segregation Distorter gene complex of Drosophila melanogaster
- PMID: 22964836
- PMCID: PMC3430544
- DOI: 10.1534/genetics.112.141390
The selfish Segregation Distorter gene complex of Drosophila melanogaster
Abstract
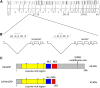
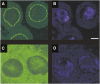
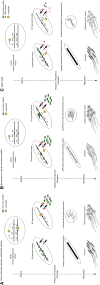
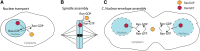
Segregation Distorter (SD) is an autosomal meiotic drive gene complex found worldwide in natural populations of Drosophila melanogaster. During spermatogenesis, SD induces dysfunction of SD(+) spermatids so that SD/SD(+) males sire almost exclusively SD-bearing progeny rather than the expected 1:1 Mendelian ratio. SD is thus evolutionarily "selfish," enhancing its own transmission at the expense of its bearers. Here we review the molecular and evolutionary genetics of SD. Genetic analyses show that the SD is a multilocus gene complex involving two key loci--the driver, Segregation distorter (Sd), and the target of drive, Responder (Rsp)--and at least three upward modifiers of distortion. Molecular analyses show that Sd encodes a truncated duplication of the gene RanGAP, whereas Rsp is a large pericentromeric block of satellite DNA. The Sd-RanGAP protein is enzymatically wild type but mislocalized within cells and, for reasons that remain unclear, appears to disrupt the histone-to-protamine transition in drive-sensitive spermatids bearing many Rsp satellite repeats but not drive-insensitive spermatids bearing few or no Rsp satellite repeats. Evolutionary analyses show that the Sd-RanGAP duplication arose recently within the D. melanogaster lineage, exploiting the preexisting and considerably older Rsp satellite locus. Once established, the SD haplotype collected enhancers of distortion and suppressors of recombination. Further dissection of the molecular genetic and cellular basis of SD-mediated distortion seems likely to provide insights into several important areas currently understudied, including the genetic control of spermatogenesis, the maintenance and evolution of satellite DNAs, the possible roles of small interfering RNAs in the germline, and the molecular population genetics of the interaction of genetic linkage and natural selection.
Figures




References
-
- Aravin A. A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., et al. , 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337–350 - PubMed
-
- Baudry E., Viginier B., Veuille M., 2004. Non-African populations of Drosophila melanogaster have a unique origin. Mol. Biol. Evol. 21: 1482–1491 - PubMed
-
- Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., et al. , 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

