Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting
- PMID: 22964890
- PMCID: PMC3530684
- DOI: 10.1101/gr.142661.112
Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting
Abstract
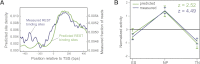
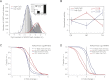
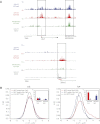
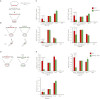
Although changes in chromatin are integral to transcriptional reprogramming during cellular differentiation, it is currently unclear how chromatin modifications are targeted to specific loci. To systematically identify transcription factors (TFs) that can direct chromatin changes during cell fate decisions, we model the relationship between genome-wide dynamics of chromatin marks and the local occurrence of computationally predicted TF binding sites. By applying this computational approach to a time course of Polycomb-mediated H3K27me3 marks during neuronal differentiation of murine stem cells, we identify several motifs that likely regulate the dynamics of this chromatin mark. Among these, the sites bound by REST and by the SNAIL family of TFs are predicted to transiently recruit H3K27me3 in neuronal progenitors. We validate these predictions experimentally and show that absence of REST indeed causes loss of H3K27me3 at target promoters in trans, specifically at the neuronal progenitor state. Moreover, using targeted transgenic insertion, we show that promoter fragments containing REST or SNAIL binding sites are sufficient to recruit H3K27me3 in cis, while deletion of these sites results in loss of H3K27me3. These findings illustrate that the occurrence of TF binding sites can determine chromatin dynamics. Local determination of Polycomb activity by REST and SNAIL motifs exemplifies such TF based regulation of chromatin. Furthermore, our results show that key TFs can be identified ab initio through computational modeling of epigenome data sets using a modeling approach that we make readily accessible.
Figures

Similar articles
-
Genome-wide recruitment to Polycomb-modified chromatin and activity regulation of the synovial sarcoma oncogene SYT-SSX2.BMC Genomics. 2012 May 17;13:189. doi: 10.1186/1471-2164-13-189. BMC Genomics. 2012. PMID: 22594313 Free PMC article.
-
Delayed Accumulation of H3K27me3 on Nascent DNA Is Essential for Recruitment of Transcription Factors at Early Stages of Stem Cell Differentiation.Mol Cell. 2017 Apr 20;66(2):247-257.e5. doi: 10.1016/j.molcel.2017.03.006. Epub 2017 Apr 11. Mol Cell. 2017. PMID: 28410996 Free PMC article.
-
Role of chromatin and transcriptional co-regulators in mediating p63-genome interactions in keratinocytes.BMC Genomics. 2014 Nov 29;15(1):1042. doi: 10.1186/1471-2164-15-1042. BMC Genomics. 2014. PMID: 25433490 Free PMC article.
-
Cell Fate and Developmental Regulation Dynamics by Polycomb Proteins and 3D Genome Architecture.Bioessays. 2019 Mar;41(3):e1800222. doi: 10.1002/bies.201800222. Epub 2019 Feb 22. Bioessays. 2019. PMID: 30793782 Review.
-
The Yin and Yang of Chromatin Dynamics In Stem Cell Fate Selection.Trends Genet. 2016 Feb;32(2):89-100. doi: 10.1016/j.tig.2015.11.002. Epub 2015 Dec 13. Trends Genet. 2016. PMID: 26689127 Free PMC article. Review.
Cited by
-
The PRC2-binding long non-coding RNAs in human and mouse genomes are associated with predictive sequence features.Sci Rep. 2017 Jan 31;7:41669. doi: 10.1038/srep41669. Sci Rep. 2017. PMID: 28139710 Free PMC article.
-
Epigenomics of Neural Cells: REST-Induced Down- and Upregulation of Gene Expression in a Two-Clone PC12 Cell Model.Biomed Res Int. 2015;2015:202914. doi: 10.1155/2015/202914. Epub 2015 Aug 27. Biomed Res Int. 2015. PMID: 26413508 Free PMC article.
-
CG dinucleotides enhance promoter activity independent of DNA methylation.Genome Res. 2019 Apr;29(4):554-563. doi: 10.1101/gr.241653.118. Epub 2019 Feb 1. Genome Res. 2019. PMID: 30709850 Free PMC article.
-
H3K27 methylation: a promiscuous repressive chromatin mark.Curr Opin Genet Dev. 2017 Apr;43:31-37. doi: 10.1016/j.gde.2016.11.001. Epub 2016 Dec 8. Curr Opin Genet Dev. 2017. PMID: 27940208 Free PMC article. Review.
-
Transcription factor binding predicts histone modifications in human cell lines.Proc Natl Acad Sci U S A. 2014 Sep 16;111(37):13367-72. doi: 10.1073/pnas.1412081111. Epub 2014 Sep 3. Proc Natl Acad Sci U S A. 2014. PMID: 25187560 Free PMC article.
References
-
- Abbott A 2011. Europe to map the human epigenome. Nature 477: 518 doi: 10.1038/477518a - PubMed
-
- Arnold P, Erb I, Pachkov M, Molina N, van Nimwegen E 2012. MotEvo: Integrated Bayesian probabilistic methods for inferring regulatory sites and motifs on multiple alignments of DNA sequences. Bioinformatics 28: 487–494 - PubMed
-
- Barrera LO, Ren B 2006. The transcriptional regulatory code of eukaryotic cells—insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr Opin Cell Biol 18: 291–298 - PubMed
-
- Beer MA, Tavazoie S 2004. Predicting gene expression from sequence. Cell 117: 185–198 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
