Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells
- PMID: 22966001
- PMCID: PMC3457733
- DOI: 10.1084/jem.20120914
Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells
Abstract
Foxp3 activity is essential for the normal function of the immune system. Two types of regulatory T (T reg) cells express Foxp3, thymus-generated natural T reg (nT reg) cells, and peripherally generated adaptive T reg (iT reg) cells. These cell types have complementary functions. Until now, it has not been possible to distinguish iT reg from nT reg cells in vivo based solely on surface markers. We report here that Neuropilin 1 (Nrp1) is expressed at high levels by most nT reg cells; in contrast, mucosa-generated iT reg and other noninflammatory iT reg cells express low levels of Nrp1. We found that Nrp1 expression is under the control of TGF-β. By tracing nT reg and iT reg cells, we could establish that some tumors have a very large proportion of infiltrating iT reg cells. iT reg cells obtained from highly inflammatory environments, such as the spinal cords of mice with spontaneous autoimmune encephalomyelitis (EAE) and the lungs of mice with chronic asthma, express Nrp1. In the same animals, iT reg cells in secondary lymphoid organs remain Nrp1(low). We also determined that, in spontaneous EAE, iT reg cells help to establish a chronic phase of the disease.
Figures





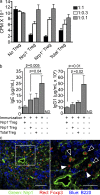
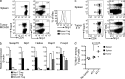



References
-
- Bennett C.L., Christie J., Ramsdell F., Brunkow M.E., Ferguson P.J., Whitesell L., Kelly T.E., Saulsbury F.T., Chance P.F., Ochs H.D. 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27:20–21 10.1038/83713 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

