Upstream stimulatory factor 2 and hypoxia-inducible factor 2α (HIF2α) cooperatively activate HIF2 target genes during hypoxia
- PMID: 22966206
- PMCID: PMC3486188
- DOI: 10.1128/MCB.00724-12
Upstream stimulatory factor 2 and hypoxia-inducible factor 2α (HIF2α) cooperatively activate HIF2 target genes during hypoxia
Abstract
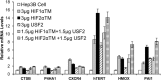
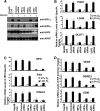
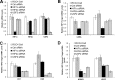
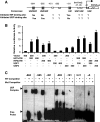
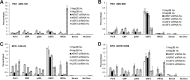
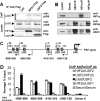
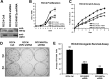
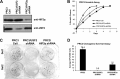
While the functions of hypoxia-inducible factor 1α (HIF1α)/aryl hydrocarbon receptor nuclear translocator (ARNT) and HIF2α/ARNT (HIF2) proteins in activating hypoxia-inducible genes are well established, the role of other transcription factors in the hypoxic transcriptional response is less clear. We report here for the first time that the basic helix-loop-helix-leucine-zip transcription factor upstream stimulatory factor 2 (USF2) is required for the hypoxic transcriptional response, specifically, for hypoxic activation of HIF2 target genes. We show that inhibiting USF2 activity greatly reduces hypoxic induction of HIF2 target genes in cell lines that have USF2 activity, while inducing USF2 activity in cells lacking USF2 activity restores hypoxic induction of HIF2 target genes. Mechanistically, USF2 activates HIF2 target genes by binding to HIF2 target gene promoters, interacting with HIF2α protein, and recruiting coactivators CBP and p300 to form enhanceosome complexes that contain HIF2α, USF2, CBP, p300, and RNA polymerase II on HIF2 target gene promoters. Functionally, the effect of USF2 knockdown on proliferation, motility, and clonogenic survival of HIF2-dependent tumor cells in vitro is phenocopied by HIF2α knockdown, indicating that USF2 works with HIF2 to activate HIF2 target genes and to drive HIF2-depedent tumorigenesis.
Figures


Similar articles
-
STAT3 or USF2 contributes to HIF target gene specificity.PLoS One. 2013 Aug 21;8(8):e72358. doi: 10.1371/journal.pone.0072358. eCollection 2013. PLoS One. 2013. PMID: 23991099 Free PMC article.
-
STAT3 and HIF1α cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells.Oncogene. 2014 Mar 27;33(13):1670-9. doi: 10.1038/onc.2013.115. Epub 2013 Apr 22. Oncogene. 2014. PMID: 23604114 Free PMC article.
-
Enhanceosomes as integrators of hypoxia inducible factor (HIF) and other transcription factors in the hypoxic transcriptional response.Cell Signal. 2013 Sep;25(9):1895-903. doi: 10.1016/j.cellsig.2013.05.018. Epub 2013 May 21. Cell Signal. 2013. PMID: 23707522 Free PMC article. Review.
-
BRG1 and BRM chromatin-remodeling complexes regulate the hypoxia response by acting as coactivators for a subset of hypoxia-inducible transcription factor target genes.Mol Cell Biol. 2013 Oct;33(19):3849-63. doi: 10.1128/MCB.00731-13. Epub 2013 Jul 29. Mol Cell Biol. 2013. PMID: 23897427 Free PMC article.
-
Biology of hypoxia-inducible factor-2alpha in development and disease.Cell Death Differ. 2008 Apr;15(4):628-34. doi: 10.1038/cdd.2008.17. Epub 2008 Feb 15. Cell Death Differ. 2008. PMID: 18259197 Free PMC article. Review.
Cited by
-
Protein kinases as switches for the function of upstream stimulatory factors: implications for tissue injury and cancer.Front Pharmacol. 2015 Feb 18;6:3. doi: 10.3389/fphar.2015.00003. eCollection 2015. Front Pharmacol. 2015. PMID: 25741280 Free PMC article.
-
FLCN Regulates HIF2α Nuclear Import and Proliferation of Clear Cell Renal Cell Carcinoma.Front Mol Biosci. 2020 Jul 28;7:121. doi: 10.3389/fmolb.2020.00121. eCollection 2020. Front Mol Biosci. 2020. PMID: 32850947 Free PMC article.
-
Intra-articular delivery of anti-Hif-2α siRNA by chondrocyte-homing nanoparticles to prevent cartilage degeneration in arthritic mice.Gene Ther. 2015 Jun;22(6):439-48. doi: 10.1038/gt.2015.16. Epub 2015 Apr 16. Gene Ther. 2015. PMID: 25876463
-
Centella asiatica extract ameliorates deoxygenation-induced neurological dysfunction in zebrafish larvae.Open Vet J. 2024 May;14(5):1154-1160. doi: 10.5455/OVJ.2024.v14.i5.9. Epub 2024 May 31. Open Vet J. 2024. PMID: 38938421 Free PMC article.
-
Genome-wide analysis of HIF-2α chromatin binding sites under normoxia in human bronchial epithelial cells (BEAS-2B) suggests its diverse functions.Sci Rep. 2016 Jul 4;6:29311. doi: 10.1038/srep29311. Sci Rep. 2016. PMID: 27373565 Free PMC article.
References
-
- Allen RR, Qi L, Higgins PJ. 2005. Upstream stimulatory factor regulates E box-dependent PAI-1 transcription in human epidermal keratinocytes. J. Cell. Physiol. 203:156–165 - PubMed
-
- Anderson CJ, Hoare SF, Ashcroft M, Bilsland AE, Keith WN. 2006. Hypoxic regulation of telomerase gene expression by transcriptional and post-transcriptional mechanisms. Oncogene 25:61–69 - PubMed
-
- Aprelikova O, Wood M, Tackett S, Chandramouli GV, Barrett JC. 2006. Role of ETS transcription factors in the hypoxia-inducible factor-2 target gene selection. Cancer Res. 66:5641–5647 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
