Guidelines for the use and interpretation of assays for monitoring autophagy
- PMID: 22966490
- PMCID: PMC3404883
- DOI: 10.4161/auto.19496
Guidelines for the use and interpretation of assays for monitoring autophagy
Abstract
In 2008 we published the first set of guidelines for standardizing research in autophagy. Since then, research on this topic has continued to accelerate, and many new scientists have entered the field. Our knowledge base and relevant new technologies have also been expanding. Accordingly, it is important to update these guidelines for monitoring autophagy in different organisms. Various reviews have described the range of assays that have been used for this purpose. Nevertheless, there continues to be confusion regarding acceptable methods to measure autophagy, especially in multicellular eukaryotes. A key point that needs to be emphasized is that there is a difference between measurements that monitor the numbers or volume of autophagic elements (e.g., autophagosomes or autolysosomes) at any stage of the autophagic process vs. those that measure flux through the autophagy pathway (i.e., the complete process); thus, a block in macroautophagy that results in autophagosome accumulation needs to be differentiated from stimuli that result in increased autophagic activity, defined as increased autophagy induction coupled with increased delivery to, and degradation within, lysosomes (in most higher eukaryotes and some protists such as Dictyostelium) or the vacuole (in plants and fungi). In other words, it is especially important that investigators new to the field understand that the appearance of more autophagosomes does not necessarily equate with more autophagy. In fact, in many cases, autophagosomes accumulate because of a block in trafficking to lysosomes without a concomitant change in autophagosome biogenesis, whereas an increase in autolysosomes may reflect a reduction in degradative activity. Here, we present a set of guidelines for the selection and interpretation of methods for use by investigators who aim to examine macroautophagy and related processes, as well as for reviewers who need to provide realistic and reasonable critiques of papers that are focused on these processes. These guidelines are not meant to be a formulaic set of rules, because the appropriate assays depend in part on the question being asked and the system being used. In addition, we emphasize that no individual assay is guaranteed to be the most appropriate one in every situation, and we strongly recommend the use of multiple assays to monitor autophagy. In these guidelines, we consider these various methods of assessing autophagy and what information can, or cannot, be obtained from them. Finally, by discussing the merits and limits of particular autophagy assays, we hope to encourage technical innovation in the field.
Figures
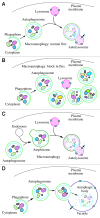
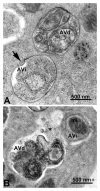

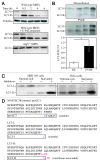
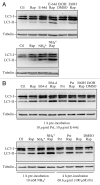
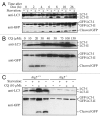
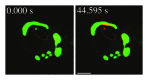
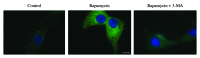
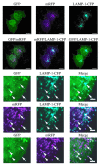
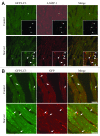
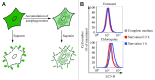
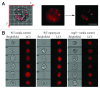





Comment in
-
The role of the Eph/ephrin-system in atherosclerotic plaque development: a complex puzzle.Cardiovasc Pathol. 2014 Jul-Aug;23(4):251. doi: 10.1016/j.carpath.2014.03.005. Epub 2014 Mar 27. Cardiovasc Pathol. 2014. PMID: 24746710 No abstract available.
References
-
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 CA140964/CA/NCI NIH HHS/United States
- G-0905/PUK_/Parkinson's UK/United Kingdom
- K05 CA142885/CA/NCI NIH HHS/United States
- R01 DK082437/DK/NIDDK NIH HHS/United States
- G0600251/MRC_/Medical Research Council/United Kingdom
- R01 HL096625/HL/NHLBI NIH HHS/United States
- R01 HL102738/HL/NHLBI NIH HHS/United States
- R01 GM099040/GM/NIGMS NIH HHS/United States
- R01 DK044234/DK/NIDDK NIH HHS/United States
- UL1 TR000005/TR/NCATS NIH HHS/United States
- R01 CA159314/CA/NCI NIH HHS/United States
- R01 DK124709/DK/NIDDK NIH HHS/United States
- R01 CA137260/CA/NCI NIH HHS/United States
- R01 EY019320/EY/NEI NIH HHS/United States
- R01 CA133228/CA/NCI NIH HHS/United States
- K01 DK087776/DK/NIDDK NIH HHS/United States
- I01 BX001969/BX/BLRD VA/United States
- R01 CA154835/CA/NCI NIH HHS/United States
- 12103/CRUK_/Cancer Research UK/United Kingdom
- 15816/CRUK_/Cancer Research UK/United Kingdom
- R01 AI042999/AI/NIAID NIH HHS/United States
- R01 DK100644/DK/NIDDK NIH HHS/United States
- BB/F012861/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 CA103632/CA/NCI NIH HHS/United States
- R01 AI064705/AI/NIAID NIH HHS/United States
- 11359/CRUK_/Cancer Research UK/United Kingdom
- K02 AG042095/AG/NIA NIH HHS/United States
- R01 CA131188/CA/NCI NIH HHS/United States
- G0700127/MRC_/Medical Research Council/United Kingdom
- R01 DK079879/DK/NIDDK NIH HHS/United States
- P01 CA108671/CA/NCI NIH HHS/United States
- R01 GM097355/GM/NIGMS NIH HHS/United States
- R01 AI089716/AI/NIAID NIH HHS/United States
- BBS/E/B/00001221/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- K08 CA164047/CA/NCI NIH HHS/United States
- R01 HL079669/HL/NHLBI NIH HHS/United States
- BBS/E/F/00042262/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 17655/ARC_/Arthritis Research UK/United Kingdom
- R01 AI080703/AI/NIAID NIH HHS/United States
- R01 DK090115/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM082830/GM/NIGMS NIH HHS/United States
- MC_U132670601/MRC_/Medical Research Council/United Kingdom
- R01 HL060590/HL/NHLBI NIH HHS/United States
- R01 CA123350/CA/NCI NIH HHS/United States
- R01 NS064090/NS/NINDS NIH HHS/United States
- R01 AI081884/AI/NIAID NIH HHS/United States
- 17285/VAC_/Versus Arthritis/United Kingdom
- 15153/CRUK_/Cancer Research UK/United Kingdom
- R01 AT005076/AT/NCCIH NIH HHS/United States
- G12 MD007599/MD/NIMHD NIH HHS/United States
- R01 GM053396/GM/NIGMS NIH HHS/United States
- R01 AG039628/AG/NIA NIH HHS/United States
- G0601840/MRC_/Medical Research Council/United Kingdom
- R01 GM062509/GM/NIGMS NIH HHS/United States
- MC_U132670600/MRC_/Medical Research Council/United Kingdom
- R01 EY009083/EY/NEI NIH HHS/United States
- R01 DK074778/DK/NIDDK NIH HHS/United States
- G12 MD007595/MD/NIMHD NIH HHS/United States
- 087518/WT_/Wellcome Trust/United Kingdom
- R01 HL085629/HL/NHLBI NIH HHS/United States
- 19379/ARC_/Arthritis Research UK/United Kingdom
- P30 AG028740/AG/NIA NIH HHS/United States
- I01 BX001318/BX/BLRD VA/United States
- MC_U142684175/MRC_/Medical Research Council/United Kingdom
- R01 CA138641/CA/NCI NIH HHS/United States
- S10 RR025596/RR/NCRR NIH HHS/United States
- R01 CA157490/CA/NCI NIH HHS/United States
- P20 GM121176/GM/NIGMS NIH HHS/United States
- R01 NS043466/NS/NINDS NIH HHS/United States
- R37 AA020518/AA/NIAAA NIH HHS/United States
- G0700949/MRC_/Medical Research Council/United Kingdom
- MC_U105170648/MRC_/Medical Research Council/United Kingdom
- G0600782/MRC_/Medical Research Council/United Kingdom
- R01 CA109182/CA/NCI NIH HHS/United States
- R01 NS046489/NS/NINDS NIH HHS/United States
- R37 AI042999/AI/NIAID NIH HHS/United States
- R01 CA143811/CA/NCI NIH HHS/United States
- R01 CA102184/CA/NCI NIH HHS/United States
- R01 AG011833/AG/NIA NIH HHS/United States
- R01 CA100857/CA/NCI NIH HHS/United States
- MC_U142684172/MRC_/Medical Research Council/United Kingdom
- G0000508/MRC_/Medical Research Council/United Kingdom
- 102974/WT_/Wellcome Trust/United Kingdom
- R01 MH073490/MH/NIMH NIH HHS/United States
- R01 NS046451/NS/NINDS NIH HHS/United States
- R01 DK073336/DK/NIDDK NIH HHS/United States
- R01 EY020491/EY/NEI NIH HHS/United States
- BB/E018521/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- J-0901/PUK_/Parkinson's UK/United Kingdom
- R01 AA019730/AA/NIAAA NIH HHS/United States
- R01 HL067724/HL/NHLBI NIH HHS/United States
- R01 GM079431/GM/NIGMS NIH HHS/United States
- R01 AI057831/AI/NIAID NIH HHS/United States
- R01 NS050396/NS/NINDS NIH HHS/United States
- R21 AG039799/AG/NIA NIH HHS/United States
- R01 DE010742/DE/NIDCR NIH HHS/United States
- 12825/CRUK_/Cancer Research UK/United Kingdom
- BB/G021422/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 CA141703/CA/NCI NIH HHS/United States
- R01 GM089919/GM/NIGMS NIH HHS/United States
- R01 CA134530/CA/NCI NIH HHS/United States
- G1002610/MRC_/Medical Research Council/United Kingdom
- TL4 MD009637/MD/NIMHD NIH HHS/United States
- R01 HL114858/HL/NHLBI NIH HHS/United States
- R01 CA142862/CA/NCI NIH HHS/United States
- R01 HL112330/HL/NHLBI NIH HHS/United States
- R01 AG033283/AG/NIA NIH HHS/United States
- R01 HL072166/HL/NHLBI NIH HHS/United States
- GM53396/GM/NIGMS NIH HHS/United States
- 095317/WT_/Wellcome Trust/United Kingdom
- R01 DK061498/DK/NIDDK NIH HHS/United States
- R01 AI079065/AI/NIAID NIH HHS/United States
- MC_UP_A500_1019/MRC_/Medical Research Council/United Kingdom
- 15890/CRUK_/Cancer Research UK/United Kingdom
- MC_U105184308/MRC_/Medical Research Council/United Kingdom
- R01 CA162405/CA/NCI NIH HHS/United States
- R37 DK050107/DK/NIDDK NIH HHS/United States
- R01 DK050610/DK/NIDDK NIH HHS/United States
- R01 DK052825/DK/NIDDK NIH HHS/United States
- R01 CA129536/CA/NCI NIH HHS/United States
- R01 HL106019/HL/NHLBI NIH HHS/United States
- R01 AG031867/AG/NIA NIH HHS/United States
- P01 AI056097/AI/NIAID NIH HHS/United States
- R01 NS080108/NS/NINDS NIH HHS/United States
- R01 AG023039/AG/NIA NIH HHS/United States
- P 23490/FWF_/Austrian Science Fund FWF/Austria
- R01 DK094652/DK/NIDDK NIH HHS/United States
- R01 CA139319/CA/NCI NIH HHS/United States
- R01 CA150214/CA/NCI NIH HHS/United States
