Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells
- PMID: 22968442
- PMCID: PMC3472693
- DOI: 10.1038/msb.2012.40
Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells
Abstract
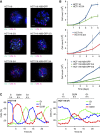
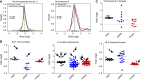
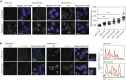
Extra chromosome copies markedly alter the physiology of eukaryotic cells, but the underlying reasons are not well understood. We created human trisomic and tetrasomic cell lines and determined the quantitative changes in their transcriptome and proteome in comparison with their diploid counterparts. We found that whereas transcription levels reflect the chromosome copy number changes, the abundance of some proteins, such as subunits of protein complexes and protein kinases, is reduced toward diploid levels. Furthermore, using the quantitative data we investigated the changes of cellular pathways in response to aneuploidy. This analysis revealed specific and uniform alterations in pathway regulation in cells with extra chromosomes. For example, the DNA and RNA metabolism pathways were downregulated, whereas several pathways such as energy metabolism, membrane metabolism and lysosomal pathways were upregulated. In particular, we found that the p62-dependent selective autophagy is activated in the human trisomic and tetrasomic cells. Our data present the first broad proteomic analysis of human cells with abnormal karyotypes and suggest a uniform cellular response to the presence of an extra chromosome.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Aït Yahya-Graison E, Aubert J, Dauphinot L, Rivals I, Prieur M, Golfier G, Rossier J, Personnaz L, Créau N, Bléhaut H, Robin S, Delabar JM, Potier MC (2007) Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am J Hum Genet 81: 475–491 - PMC - PubMed
-
- Birchler JA, Riddle NC, Auger DL, Veitia RA (2005) Dosage balance in gene regulation: biological implications. Trends Genet 21: 219–226 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

