Regulating type 1 IFN effects in CD8 T cells during viral infections: changing STAT4 and STAT1 expression for function
- PMID: 22968462
- PMCID: PMC3488885
- DOI: 10.1182/blood-2012-05-428672
Regulating type 1 IFN effects in CD8 T cells during viral infections: changing STAT4 and STAT1 expression for function
Abstract
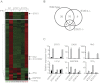
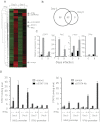
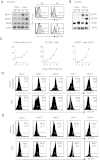
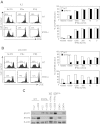
Type 1 IFNs can conditionally activate all of the signal transducers and activators of transcription molecules (STATs), including STAT4. The best-characterized signaling pathways use STAT1, however, and type 1 IFN inhibition of cell proliferation is STAT1 dependent. We report that type 1 IFNs can basally stimulate STAT1- and STAT4-dependent effects in CD8 T cells, but that CD8 T cells responding to infections of mice with lymphocytic choriomenigitis virus have elevated STAT4 and lower STAT1 expression with significant consequences for modifying the effects of type 1 IFN exposure. The phenotype was associated with preferential type 1 IFN activation of STAT4 compared with STAT1. Stimulation through the TCR induced elevated STAT4 expression, and STAT4 was required for peak expansion of antigen-specific CD8 T cells, low STAT1 levels, and resistance to type 1 IFN-mediated inhibition of proliferation. Thus, a mechanism is discovered for regulating the consequences of type 1 IFN exposure in CD8 T cells, with STAT4 acting as a key molecule in driving optimal antigen-specific responses and overcoming STAT1-dependent inhibition of proliferation.
Figures


Similar articles
-
High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells.J Exp Med. 2007 Oct 1;204(10):2383-96. doi: 10.1084/jem.20070401. Epub 2007 Sep 10. J Exp Med. 2007. PMID: 17846149 Free PMC article.
-
CD8 T cells in innate immune responses: using STAT4-dependent but antigen-independent pathways to gamma interferon during viral infection.mBio. 2014 Oct 21;5(5):e01978-14. doi: 10.1128/mBio.01978-14. mBio. 2014. PMID: 25336459 Free PMC article.
-
Type 1 interferon induction of natural killer cell gamma interferon production for defense during lymphocytic choriomeningitis virus infection.mBio. 2011 Aug 9;2(4):e00169-11. doi: 10.1128/mBio.00169-11. Print 2011. mBio. 2011. PMID: 21828218 Free PMC article.
-
T helper differentiation proceeds through Stat1-dependent, Stat4-dependent and Stat4-independent phases.Curr Top Microbiol Immunol. 1999;238:13-26. doi: 10.1007/978-3-662-09709-0_2. Curr Top Microbiol Immunol. 1999. PMID: 10087648 Review.
-
NK cells and interferons.Cytokine Growth Factor Rev. 2015 Apr;26(2):113-20. doi: 10.1016/j.cytogfr.2014.11.003. Epub 2014 Nov 13. Cytokine Growth Factor Rev. 2015. PMID: 25443799 Review.
Cited by
-
Aberrant signaling of immune cells in Sjögren's syndrome patient subgroups upon interferon stimulation.Front Immunol. 2022 Aug 22;13:854183. doi: 10.3389/fimmu.2022.854183. eCollection 2022. Front Immunol. 2022. PMID: 36072585 Free PMC article.
-
Influenza-induced Tpl2 expression within alveolar epithelial cells is dispensable for host viral control and anti-viral immunity.PLoS One. 2022 Jan 20;17(1):e0262832. doi: 10.1371/journal.pone.0262832. eCollection 2022. PLoS One. 2022. PMID: 35051238 Free PMC article.
-
Interferon-Driven Immune Dysregulation in Down Syndrome: A Review of the Evidence.J Inflamm Res. 2021 Oct 7;14:5187-5200. doi: 10.2147/JIR.S280953. eCollection 2021. J Inflamm Res. 2021. PMID: 34675597 Free PMC article. Review.
-
Opposing Roles of Type I Interferons in Cancer Immunity.Annu Rev Pathol. 2021 Jan 24;16:167-198. doi: 10.1146/annurev-pathol-031920-093932. Epub 2020 Dec 2. Annu Rev Pathol. 2021. PMID: 33264572 Free PMC article. Review.
-
Memory CD8+ T Cell Protection From Viral Reinfection Depends on Interleukin-33 Alarmin Signals.Front Immunol. 2019 Aug 7;10:1833. doi: 10.3389/fimmu.2019.01833. eCollection 2019. Front Immunol. 2019. PMID: 31447845 Free PMC article.
References
-
- Biron CA. Interferons alpha and beta as immune regulators–a new look. Immunity. 2001;14(6):661–664. - PubMed
-
- Brierley MM, Fish EN. Review: IFN-alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J Interferon Cytokine Res. 2002;22(8):835–845. - PubMed
-
- García-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312(5775):879–882. - PubMed
-
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

