A microfluidic platform for rapid, stress-induced antibiotic susceptibility testing of Staphylococcus aureus
- PMID: 22968495
- PMCID: PMC3489182
- DOI: 10.1039/c2lc40531h
A microfluidic platform for rapid, stress-induced antibiotic susceptibility testing of Staphylococcus aureus
Erratum in
- Lab Chip. 2013 Dec 21;13(24):4890
Abstract
The emergence and spread of bacterial resistance to ever increasing classes of antibiotics intensifies the need for fast phenotype-based clinical tests for determining antibiotic susceptibility. Standard susceptibility testing relies on the passive observation of bacterial growth inhibition in the presence of antibiotics. In this paper, we present a novel microfluidic platform for antibiotic susceptibility testing based on stress-activation of biosynthetic pathways that are the primary targets of antibiotics. We chose Staphylococcus aureus (S. aureus) as a model system due to its clinical importance, and we selected bacterial cell wall biosynthesis as the primary target of both stress and antibiotic. Enzymatic and mechanical stresses were used to damage the bacterial cell wall, and a β-lactam antibiotic interfered with the repair process, resulting in rapid cell death of strains that harbor no resistance mechanism. In contrast, resistant bacteria remained viable under the assay conditions. Bacteria, covalently-bound to the bottom of the microfluidic channel, were subjected to mechanical shear stress created by flowing culture media through the microfluidic channel and to enzymatic stress with sub-inhibitory concentrations of the bactericidal agent lysostaphin. Bacterial cell death was monitored via fluorescence using the Sytox Green dead cell stain, and rates of killing were measured for the bacterial samples in the presence and absence of oxacillin. Using model susceptible (Sanger 476) and resistant (MW2) S. aureus strains, a metric was established to separate susceptible and resistant staphylococci based on normalized fluorescence values after 60 min of exposure to stress and antibiotic. Because this ground-breaking approach is not based on standard methodology, it circumvents the need for minimum inhibitory concentration (MIC) measurements and long wait times. We demonstrate the successful development of a rapid microfluidic-based and stress-activated antibiotic susceptibility test by correctly designating the phenotypes of 16 additional clinically relevant S. aureus strains in a blinded study. In addition to future clinical utility, this method has great potential for studying the effects of various stresses on bacteria and their antibiotic susceptibility.
Figures


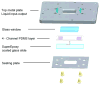

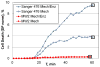


Similar articles
-
Stress-induced antibiotic susceptibility testing on a chip.J Vis Exp. 2014 Jan 8;(83):e50828. doi: 10.3791/50828. J Vis Exp. 2014. PMID: 24430495 Free PMC article.
-
Ultrafast Parallelized Microfluidic Platform for Antimicrobial Susceptibility Testing of Gram Positive and Negative Bacteria.Anal Chem. 2019 May 7;91(9):6242-6249. doi: 10.1021/acs.analchem.9b00939. Epub 2019 Apr 15. Anal Chem. 2019. PMID: 30938989
-
Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system.Lab Chip. 2013 Jan 21;13(2):280-7. doi: 10.1039/c2lc41055a. Epub 2012 Nov 21. Lab Chip. 2013. PMID: 23172338
-
Label-free single-cell antimicrobial susceptibility testing in droplets with concentration gradient generation.Lab Chip. 2024 Dec 3;24(24):5274-5289. doi: 10.1039/d4lc00629a. Lab Chip. 2024. PMID: 39324417
-
Methods for Rapid Evaluation of Microbial Antibiotics Resistance.Biochemistry (Mosc). 2025 Jan;90(Suppl 1):S312-S341. doi: 10.1134/S0006297924603678. Biochemistry (Mosc). 2025. PMID: 40164164 Review.
Cited by
-
A Continuous Microfluidic Concentrator for High-Sensitivity Detection of Bacteria in Water Sources.Micromachines (Basel). 2022 Jul 10;13(7):1093. doi: 10.3390/mi13071093. Micromachines (Basel). 2022. PMID: 35888910 Free PMC article.
-
Next-generation rapid phenotypic antimicrobial susceptibility testing.Nat Commun. 2024 Nov 9;15(1):9719. doi: 10.1038/s41467-024-53930-x. Nat Commun. 2024. PMID: 39521792 Free PMC article. Review.
-
Rapid antibiotic sensitivity testing in microwell arrays.Technology (Singap World Sci). 2017 Jun;5(2):107-114. doi: 10.1142/S2339547817500030. Epub 2017 May 16. Technology (Singap World Sci). 2017. PMID: 28781994 Free PMC article.
-
A Prospective Evaluation of Two Rapid Phenotypical Antimicrobial Susceptibility Technologies for the Diagnostic Stewardship of Sepsis.Biomed Res Int. 2018 May 10;2018:6976923. doi: 10.1155/2018/6976923. eCollection 2018. Biomed Res Int. 2018. PMID: 29862284 Free PMC article.
-
A general method for rapid determination of antibiotic susceptibility and species in bacterial infections.J Clin Microbiol. 2015 Feb;53(2):425-32. doi: 10.1128/JCM.02434-14. Epub 2014 Nov 19. J Clin Microbiol. 2015. PMID: 25411178 Free PMC article.
References
-
- Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK and the Active Bacterial Core surveillance (ABCs) MRSA Investigators, . JAMA. 2007;298:1763–1771. - PubMed
-
- Klevens RM, Edwards JR, Tenover FC, McDonald LC, Horan T, Gaynes R and the National Nosocomial Infections Surveillance System. Clin Infect Dis. 2006;42:389–391. - PubMed
-
- Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, Shah S, Rudrik JT, Pupp GR, Brown WJ, Cardo D, Fridkin SK and the Vancomycin-Resistant Staphylococcus aureus Investigative Team, . N Engl J Med. 2003;348:1342–1347. - PubMed
-
- Sanchez Garcia M, De la Torre MA, Morales G, Pelaez B, Tolon MJ, Domingo S, Candel FJ, Andrade R, Arribi A, Garcia N, Martinez Sagasti F, Fereres J, Picazo J. JAMA. 2010;303:2260–2264. - PubMed
-
- Davey F, Henry JB, Herman CJ, editors. Clinical diagnosis and management by laboratory methods. 20. W.B. Saunders; Philadelphia PA: 2001.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

