DNA methylation at differentially methylated regions of imprinted genes is resistant to developmental programming by maternal nutrition
- PMID: 22968513
- PMCID: PMC3469461
- DOI: 10.4161/epi.22141
DNA methylation at differentially methylated regions of imprinted genes is resistant to developmental programming by maternal nutrition
Abstract
The nutritional environment in which the mammalian fetus or infant develop is recognized as influencing the risk of chronic diseases, such as type 2 diabetes and hypertension, in a phenomenon that has become known as developmental programming. The late onset of such diseases in response to earlier transient experiences has led to the suggestion that developmental programming may have an epigenetic component, because epigenetic marks such as DNA methylation or histone tail modifications could provide a persistent memory of earlier nutritional states. One class of genes that has been considered a potential target or mediator of programming events is imprinted genes, because these genes critically depend upon epigenetic modifications for correct expression and because many imprinted genes have roles in controlling fetal growth as well as neonatal and adult metabolism. In this study, we have used an established model of developmental programming-isocaloric protein restriction to female mice during gestation or lactation-to examine whether there are effects on expression and DNA methylation of imprinted genes in the offspring. We find that although expression of some imprinted genes in liver of offspring is robustly and sustainably changed, methylation of the differentially methylated regions (DMRs) that control their monoallelic expression remains largely unaltered. We conclude that deregulation of imprinting through a general effect on DMR methylation is unlikely to be a common factor in developmental programming.
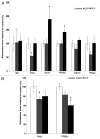
Figures



Similar articles
-
In-utero stress and mode of conception: impact on regulation of imprinted genes, fetal development and future health.Hum Reprod Update. 2019 Nov 5;25(6):777-801. doi: 10.1093/humupd/dmz025. Hum Reprod Update. 2019. PMID: 31633761
-
The loss of imprinted DNA methylation in mouse blastocysts is inflicted to a similar extent by in vitro follicle culture and ovulation induction.Mol Hum Reprod. 2016 Jun;22(6):427-41. doi: 10.1093/molehr/gaw013. Epub 2016 Feb 7. Mol Hum Reprod. 2016. PMID: 26908643
-
Deregulation of imprinted genes expression and epigenetic regulators in placental tissue from intrauterine growth restriction.J Assist Reprod Genet. 2021 Apr;38(4):791-801. doi: 10.1007/s10815-020-02047-3. Epub 2021 Jan 3. J Assist Reprod Genet. 2021. PMID: 33389447 Free PMC article.
-
Maternal epigenetics and fetal and neonatal growth.Curr Opin Endocrinol Diabetes Obes. 2017 Feb;24(1):43-46. doi: 10.1097/MED.0000000000000305. Curr Opin Endocrinol Diabetes Obes. 2017. PMID: 27898587 Review.
-
Developmental regulation of somatic imprints.Differentiation. 2011 Jun;81(5):270-80. doi: 10.1016/j.diff.2011.01.007. Epub 2011 Feb 11. Differentiation. 2011. PMID: 21316143 Review.
Cited by
-
Differential CpG methylation at Nnat in the early establishment of beta cell heterogeneity.bioRxiv [Preprint]. 2023 Nov 30:2023.02.04.527050. doi: 10.1101/2023.02.04.527050. bioRxiv. 2023. Update in: Diabetologia. 2024 Jun;67(6):1079-1094. doi: 10.1007/s00125-024-06123-6. PMID: 38076935 Free PMC article. Updated. Preprint.
-
Visualizing Changes in Cdkn1c Expression Links Early-Life Adversity to Imprint Mis-regulation in Adults.Cell Rep. 2017 Jan 31;18(5):1090-1099. doi: 10.1016/j.celrep.2017.01.010. Cell Rep. 2017. PMID: 28147266 Free PMC article.
-
Epigenetics of gestational diabetes mellitus and offspring health: the time for action is in early stages of life.Mol Hum Reprod. 2013 Jul;19(7):415-22. doi: 10.1093/molehr/gat020. Epub 2013 Mar 20. Mol Hum Reprod. 2013. PMID: 23515667 Free PMC article.
-
Degree of methylation of ZAC1 (PLAGL1) is associated with prenatal and post-natal growth in healthy infants of the EDEN mother child cohort.Epigenetics. 2014 Mar;9(3):338-45. doi: 10.4161/epi.27387. Epub 2013 Dec 6. Epigenetics. 2014. PMID: 24316753 Free PMC article.
-
Genomic imprinting and its effects on postnatal growth and adult metabolism.Cell Mol Life Sci. 2019 Oct;76(20):4009-4021. doi: 10.1007/s00018-019-03197-z. Epub 2019 Jul 3. Cell Mol Life Sci. 2019. PMID: 31270580 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
- BB/E002161/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/B/0000C169/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- B/E002161/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- FS/09/029/27902/BHF_/British Heart Foundation/United Kingdom
- G0600717/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
