Insights from genomics into bacterial pathogen populations
- PMID: 22969423
- PMCID: PMC3435253
- DOI: 10.1371/journal.ppat.1002874
Insights from genomics into bacterial pathogen populations
Abstract
Bacterial pathogens impose a heavy burden of disease on human populations worldwide. The gravest threats are posed by highly virulent respiratory pathogens, enteric pathogens, and HIV-associated infections. Tuberculosis alone is responsible for the deaths of 1.5 million people annually. Treatment options for bacterial pathogens are being steadily eroded by the evolution and spread of drug resistance. However, population-level whole genome sequencing offers new hope in the fight against pathogenic bacteria. By providing insights into bacterial evolution and disease etiology, these approaches pave the way for novel interventions and therapeutic targets. Sequencing populations of bacteria across the whole genome provides unprecedented resolution to investigate (i) within-host evolution, (ii) transmission history, and (iii) population structure. Moreover, advances in rapid benchtop sequencing herald a new era of real-time genomics in which sequencing and analysis can be deployed within hours in response to rapidly changing public health emergencies. The purpose of this review is to highlight the transformative effect of population genomics on bacteriology, and to consider the prospects for answering abiding questions such as why bacteria cause disease.
Conflict of interest statement
The author has declared that no competing interests exist.
Figures
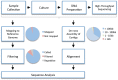
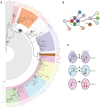

References
-
- World Health Organization (2008) The global burden of disease: 2004 update. Available: http://www.who.int/healthinfo/global_burden_disease. Accessed 10 August 2012.
-
- World Health Organization (2012) Global invasive bacterial vaccine preventable diseases (IB-VPD) information and surveillance bulletin. Volume 5. Available: http://www.who.int/nuvi/surveillance/resources/en/index.html. Accessed 10 August 2012.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

