BZLF1 governs CpG-methylated chromatin of Epstein-Barr Virus reversing epigenetic repression
- PMID: 22969425
- PMCID: PMC3435241
- DOI: 10.1371/journal.ppat.1002902
BZLF1 governs CpG-methylated chromatin of Epstein-Barr Virus reversing epigenetic repression
Abstract
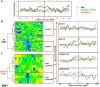
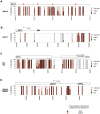
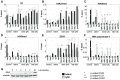
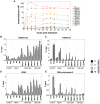
Epigenetic mechanisms are essential for the regulation of all genes in mammalian cells but transcriptional repression including DNA methylation are also major epigenetic mechanisms of defense inactivating potentially harmful pathogens. Epstein-Barr Virus (EBV), however, has evolved to take advantage of CpG methylated DNA to regulate its own biphasic life cycle. We show here that latent EBV DNA has an extreme composition of methylated CpG dinucleotides with a bimodal distribution of unmethylated or fully methylated DNA at active latent genes or completely repressed lytic promoters, respectively. We find this scenario confirmed in primary EBV-infected memory B cells in vivo. Extensive CpG methylation of EBV's DNA argues for a very restricted gene expression during latency. Above-average nucleosomal occupancy, repressive histone marks, and Polycomb-mediated epigenetic silencing further shield early lytic promoters from activation during latency. The very tight repression of viral lytic genes must be overcome when latent EBV enters its lytic phase and supports de novo virus synthesis in infected cells. The EBV-encoded and AP-1 related transcription factor BZLF1 overturns latency and initiates virus synthesis in latently infected cells. Paradoxically, BZLF1 preferentially binds to CpG-methylated motifs in key viral promoters for their activation. Upon BZLF1 binding, we find nucleosomes removed, Polycomb repression lost, and RNA polymerase II recruited to the activated early promoters promoting efficient lytic viral gene expression. Surprisingly, DNA methylation is maintained throughout this phase of viral reactivation and is no hindrance to active transcription of extensively CpG methylated viral genes as thought previously. Thus, we identify BZLF1 as a pioneer factor that reverses epigenetic silencing of viral DNA to allow escape from latency and report on a new paradigm of gene regulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Singal R, Ginder GD (1999) DNA methylation. Blood 93: 4059–4070. - PubMed
-
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, et al. (2008) Transient cyclical methylation of promoter DNA. Nature 452: 112–115. - PubMed
-
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, et al. (2008) Cyclical DNA methylation of a transcriptionally active promoter. Nature 452: 45–50. - PubMed
-
- Reid G, Gallais R, Metivier R (2009) Marking time: the dynamic role of chromatin and covalent modification in transcription. Int J Biochem Cell Biol 41: 155–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

