Ifitm3 limits the severity of acute influenza in mice
- PMID: 22969429
- PMCID: PMC3435252
- DOI: 10.1371/journal.ppat.1002909
Ifitm3 limits the severity of acute influenza in mice
Abstract
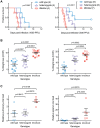
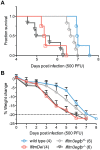
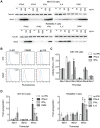
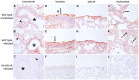
Interferon-induced transmembrane (IFITM) proteins are a family of viral restriction factors that inhibit the entry processes of several pathogenic viruses, including influenza A virus (IAV), in vitro. Here we report that IAV-infected knockout mice lacking the Ifitm locus on chromosome 7 exhibited accelerated disease progression, greater mortality, and higher pulmonary and systemic viral burdens as compared to wild type controls. We further observed that the phenotype of Ifitm3-specific knockout mice was indistinguishable from that of mice lacking the entire Ifitm locus. Ifitm3 was expressed by IAV target cells including alveolar type II pneumocytes and tracheal/bronchial respiratory epithelial cells. Robust Ifitm3 expression was also observed in several tissues in the absence of infection. Among murine Ifitm promoters, only that of Ifitm3 could be induced by type I and II interferons. Ifitm3 could also be upregulated by the gp130 cytokines IL-6 and oncostatin M on cells expressing appropriate receptors, suggesting that multiple cytokine signals could contribute to Ifitm3 expression in a cell or tissue-specific manner. Collectively, these findings establish a central role for Ifitm3 in limiting acute influenza in vivo, and provide further insight into Ifitm3 expression and regulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
- T32 RR007000/RR/NCRR NIH HHS/United States
- RR007000/RR/NCRR NIH HHS/United States
- K99AI093679/AI/NIAID NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- K99 AI093679/AI/NIAID NIH HHS/United States
- R00 AI093679/AI/NIAID NIH HHS/United States
- RR000168/RR/NCRR NIH HHS/United States
- L40AI091009/AI/NIAID NIH HHS/United States
- U54 AI057159/AI/NIAID NIH HHS/United States
- L40 AI091009/AI/NIAID NIH HHS/United States
- P51 RR000168/RR/NCRR NIH HHS/United States
- K26 RR000168/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

