Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence
- PMID: 22969433
- PMCID: PMC3435247
- DOI: 10.1371/journal.pgen.1002887
Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence
Abstract
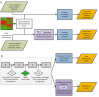
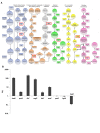
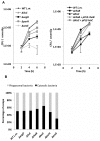
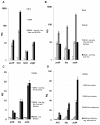
Intracellular bacterial pathogens are metabolically adapted to grow within mammalian cells. While these adaptations are fundamental to the ability to cause disease, we know little about the relationship between the pathogen's metabolism and virulence. Here we used an integrative Metabolic Analysis Tool that combines transcriptome data with genome-scale metabolic models to define the metabolic requirements of Listeria monocytogenes during infection. Twelve metabolic pathways were identified as differentially active during L. monocytogenes growth in macrophage cells. Intracellular replication requires de novo synthesis of histidine, arginine, purine, and branch chain amino acids (BCAAs), as well as catabolism of L-rhamnose and glycerol. The importance of each metabolic pathway during infection was confirmed by generation of gene knockout mutants in the respective pathways. Next, we investigated the association of these metabolic requirements in the regulation of L. monocytogenes virulence. Here we show that limiting BCAA concentrations, primarily isoleucine, results in robust induction of the master virulence activator gene, prfA, and the PrfA-regulated genes. This response was specific and required the nutrient responsive regulator CodY, which is known to bind isoleucine. Further analysis demonstrated that CodY is involved in prfA regulation, playing a role in prfA activation under limiting conditions of BCAAs. This study evidences an additional regulatory mechanism underlying L. monocytogenes virulence, placing CodY at the crossroads of metabolism and virulence.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Cross Talk between SigB and PrfA in Listeria monocytogenes Facilitates Transitions between Extra- and Intracellular Environments.Microbiol Mol Biol Rev. 2019 Sep 4;83(4):e00034-19. doi: 10.1128/MMBR.00034-19. Print 2019 Nov 20. Microbiol Mol Biol Rev. 2019. PMID: 31484692 Free PMC article. Review.
-
The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA.Mol Microbiol. 2015 Feb;95(4):624-44. doi: 10.1111/mmi.12890. Epub 2014 Dec 30. Mol Microbiol. 2015. PMID: 25430920 Free PMC article.
-
Systems Level Analyses Reveal Multiple Regulatory Activities of CodY Controlling Metabolism, Motility and Virulence in Listeria monocytogenes.PLoS Genet. 2016 Feb 19;12(2):e1005870. doi: 10.1371/journal.pgen.1005870. eCollection 2016 Feb. PLoS Genet. 2016. PMID: 26895237 Free PMC article.
-
Metabolic Genetic Screens Reveal Multidimensional Regulation of Virulence Gene Expression in Listeria monocytogenes and an Aminopeptidase That Is Critical for PrfA Protein Activation.Infect Immun. 2017 May 23;85(6):e00027-17. doi: 10.1128/IAI.00027-17. Print 2017 Jun. Infect Immun. 2017. PMID: 28396325 Free PMC article.
-
Regulation of Listeria virulence: PrfA master and commander.Curr Opin Microbiol. 2011 Apr;14(2):118-27. doi: 10.1016/j.mib.2011.01.005. Epub 2011 Mar 8. Curr Opin Microbiol. 2011. PMID: 21388862 Review.
Cited by
-
Cross Talk between SigB and PrfA in Listeria monocytogenes Facilitates Transitions between Extra- and Intracellular Environments.Microbiol Mol Biol Rev. 2019 Sep 4;83(4):e00034-19. doi: 10.1128/MMBR.00034-19. Print 2019 Nov 20. Microbiol Mol Biol Rev. 2019. PMID: 31484692 Free PMC article. Review.
-
Importance of Metabolic Adaptations in Francisella Pathogenesis.Front Cell Infect Microbiol. 2017 Mar 28;7:96. doi: 10.3389/fcimb.2017.00096. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28401066 Free PMC article. Review.
-
Regulating the Intersection of Metabolism and Pathogenesis in Gram-positive Bacteria.Microbiol Spectr. 2015 Jun;3(3):10.1128/microbiolspec.MBP-0004-2014. doi: 10.1128/microbiolspec.MBP-0004-2014. Microbiol Spectr. 2015. PMID: 26185086 Free PMC article. Review.
-
The σB-dependent regulatory sRNA Rli47 represses isoleucine biosynthesis in Listeria monocytogenes through a direct interaction with the ilvA transcript.RNA Biol. 2019 Oct;16(10):1424-1437. doi: 10.1080/15476286.2019.1632776. Epub 2019 Jun 26. RNA Biol. 2019. PMID: 31242083 Free PMC article.
-
Integrating proteomic data with metabolic modeling provides insight into key pathways of Bordetella pertussis biofilms.Front Microbiol. 2023 Aug 3;14:1169870. doi: 10.3389/fmicb.2023.1169870. eCollection 2023. Front Microbiol. 2023. PMID: 37601354 Free PMC article.
References
-
- Ray K, Marteyn B, Sansonetti PJ, Tang CM (2009) Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol 7: 333–340. - PubMed
-
- Swaminathan B, Gerner-Smidt P (2007) The epidemiology of human listeriosis. Microbes Infect 9: 1236–1243. - PubMed
-
- Bierne H, Sabet C, Personnic N, Cossart P (2007) Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect 9: 1156–1166. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

