Automated diagnoses of attention deficit hyperactive disorder using magnetic resonance imaging
- PMID: 22969709
- PMCID: PMC3431009
- DOI: 10.3389/fnsys.2012.00061
Automated diagnoses of attention deficit hyperactive disorder using magnetic resonance imaging
Abstract
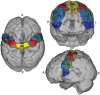
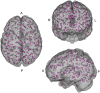
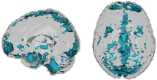
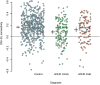
Successful automated diagnoses of attention deficit hyperactive disorder (ADHD) using imaging and functional biomarkers would have fundamental consequences on the public health impact of the disease. In this work, we show results on the predictability of ADHD using imaging biomarkers and discuss the scientific and diagnostic impacts of the research. We created a prediction model using the landmark ADHD 200 data set focusing on resting state functional connectivity (rs-fc) and structural brain imaging. We predicted ADHD status and subtype, obtained by behavioral examination, using imaging data, intelligence quotients and other covariates. The novel contributions of this manuscript include a thorough exploration of prediction and image feature extraction methodology on this form of data, including the use of singular value decompositions (SVDs), CUR decompositions, random forest, gradient boosting, bagging, voxel-based morphometry, and support vector machines as well as important insights into the value, and potentially lack thereof, of imaging biomarkers of disease. The key results include the CUR-based decomposition of the rs-fc-fMRI along with gradient boosting and the prediction algorithm based on a motor network parcellation and random forest algorithm. We conjecture that the CUR decomposition is largely diagnosing common population directions of head motion. Of note, a byproduct of this research is a potential automated method for detecting subtle in-scanner motion. The final prediction algorithm, a weighted combination of several algorithms, had an external test set specificity of 94% with sensitivity of 21%. The most promising imaging biomarker was a correlation graph from a motor network parcellation. In summary, we have undertaken a large-scale statistical exploratory prediction exercise on the unique ADHD 200 data set. The exercise produced several potential leads for future scientific exploration of the neurological basis of ADHD.
Keywords: gradient boosting; random forest; singular value decomposition; voxel-based morphometry.
Figures



Similar articles
-
Classification of drug-naive children with attention-deficit/hyperactivity disorder from typical development controls using resting-state fMRI and graph theoretical approach.Front Hum Neurosci. 2022 Aug 18;16:948706. doi: 10.3389/fnhum.2022.948706. eCollection 2022. Front Hum Neurosci. 2022. PMID: 36061501 Free PMC article.
-
Identifying individuals with attention-deficit/hyperactivity disorder based on multisite resting-state functional magnetic resonance imaging: A radiomics analysis.Hum Brain Mapp. 2023 Jun 1;44(8):3433-3445. doi: 10.1002/hbm.26290. Epub 2023 Mar 27. Hum Brain Mapp. 2023. PMID: 36971664 Free PMC article.
-
Functional Connectivity of Child and Adolescent Attention Deficit Hyperactivity Disorder Patients: Correlation with IQ.Front Hum Neurosci. 2016 Nov 9;10:565. doi: 10.3389/fnhum.2016.00565. eCollection 2016. Front Hum Neurosci. 2016. PMID: 27881961 Free PMC article.
-
Machine Learning Techniques for the Diagnosis of Attention-Deficit/Hyperactivity Disorder from Magnetic Resonance Imaging: A Concise Review.Neurol India. 2021 Nov-Dec;69(6):1518-1523. doi: 10.4103/0028-3886.333520. Neurol India. 2021. PMID: 34979636 Review.
-
A Review of the Default Mode Network in Autism Spectrum Disorders and Attention Deficit Hyperactivity Disorder.Brain Connect. 2021 May;11(4):253-263. doi: 10.1089/brain.2020.0865. Epub 2021 Feb 18. Brain Connect. 2021. PMID: 33403915 Free PMC article. Review.
Cited by
-
Neuroimaging-based biomarker discovery and validation.Pain. 2015 Aug;156(8):1379-1381. doi: 10.1097/j.pain.0000000000000223. Pain. 2015. PMID: 25970320 Free PMC article. No abstract available.
-
Classifying adolescent attention-deficit/hyperactivity disorder (ADHD) based on functional and structural imaging.Eur Child Adolesc Psychiatry. 2015 Oct;24(10):1279-89. doi: 10.1007/s00787-015-0678-4. Epub 2015 Jan 23. Eur Child Adolesc Psychiatry. 2015. PMID: 25613588
-
Intrinsic Functional Connectivity in Attention-Deficit/Hyperactivity Disorder: A Science in Development.Biol Psychiatry Cogn Neurosci Neuroimaging. 2016 May;1(3):253-261. doi: 10.1016/j.bpsc.2016.03.004. Biol Psychiatry Cogn Neurosci Neuroimaging. 2016. PMID: 27713929 Free PMC article.
-
Multivariate analyses applied to fetal, neonatal and pediatric MRI of neurodevelopmental disorders.Neuroimage Clin. 2015 Oct 3;9:532-44. doi: 10.1016/j.nicl.2015.09.017. eCollection 2015. Neuroimage Clin. 2015. PMID: 26640765 Free PMC article.
-
Neuroimaging of Small Vessel Disease in Late-Life Depression.Adv Exp Med Biol. 2019;1192:95-115. doi: 10.1007/978-981-32-9721-0_5. Adv Exp Med Biol. 2019. PMID: 31705491 Free PMC article.
References
-
- American Psychiatric Association. (2000). Diagnostic and Statistical Manual of Mental Disorders: DSM-IV-TR. American Psychiatric Publishing, Inc.
-
- Biswal B. B., Mennes M., Zuo X. N., Gohel S., Kelly C., Smith S. M., Beckmann C. F., Adelstein J. S., Buckner R. L., Colcombe S., Dogonowski A.-M., Ernst M., Fair D., Hampson M., Hoptman M. J., Hyde J. S., Kiviniemi V. J., Kötter R., Li S.-J., Lin C.-P., Lowe M. J., Mackay C., Madden D. J., Madsen K. H., Margulies D. S., Mayberg H. S., McMahon K., Monk C. S., Mostofsky S. H., Nagel B. J., Pekar J. J., Peltier S. J., Petersen S. E., Riedl V., Rombouts S. A. R. B., Rypma B., Schlaggar B. L., Schmidt S., Seidler R. D., Siegle G. J., Sorg C., Teng G.-J., Veijola J., Villringer A., Walter M., Wang L., Weng X.-C., Whitfield-Gabrieli S., Williamson P., Windischberger C., Zang Y.-F., Zhang H.-Y., Castellanos F. X., Milham M. P. (2010). Toward discovery science of human brain function. Proc. Natl. Acad. Sci. U.S.A. 1070, 4734–4739. 10.1073/pnas.0911855107 - DOI - PMC - PubMed
-
- Blei D. M., Ng A. Y., Jordan M. I. (2003). Latent dirichlet allocation. J. Mach. Learn. Res. 3, 993–1022.
-
- Breiman L. (2001). Random forests. Mach. Learn. 450, 5–32.
-
- Brett M., Christoff K., Cusack R., Lancaster J. (2004). Using the talairach atlas with the mni template. Neuroimage 130, 85.
Grants and funding
LinkOut - more resources
Full Text Sources

