Phage-Like Streptococcus pyogenes Chromosomal Islands (SpyCI) and Mutator Phenotypes: Control by Growth State and Rescue by a SpyCI-Encoded Promoter
- PMID: 22969756
- PMCID: PMC3430984
- DOI: 10.3389/fmicb.2012.00317
Phage-Like Streptococcus pyogenes Chromosomal Islands (SpyCI) and Mutator Phenotypes: Control by Growth State and Rescue by a SpyCI-Encoded Promoter
Abstract
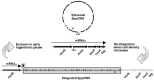
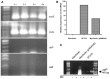
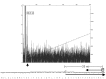
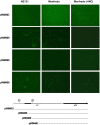
We recently showed that a prophage-like Streptococcus pyogenes chromosomal island (SpyCI) controls DNA mismatch repair and other repair functions in M1 genome strain SF370 by dynamic excision and reintegration into the 5' end of mutL in response to growth, causing the cell to alternate between a wild type and mutator phenotype. Nine of the 16 completed S. pyogenes genomes contain related SpyCI integrated into the identical attachment site in mutL, and in this study we examined a number of these strains to determine whether they also had a mutator phenotype as in SF370. With the exception of M5 genome strain Manfredo, all demonstrated a mutator phenotype as compared to SpyCI-free strain NZ131. The integrase gene (int) in the SpyCIM5 contains a deletion that rendered it inactive, and this deletion predicts that Manfredo would have a pronounced mutator phenotype. Remarkably, this was found not to be the case, but rather a cryptic promoter within the int ORF was identified that ensured constitutive expression of mutL and the downstream genes encoded on the same mRNA, providing a striking example of rescue of gene function following decay of a mobile genetic element. The frequent occurrence of SpyCI in the group A streptococci may facilitate bacterial survival by conferring an inducible mutator phenotype that promotes adaptation in the face of environmental challenges or host immunity.
Keywords: DNA mismatch repair; SpyCI; Streptococcus pyogenes; chromosomal islands; group A streptococcus; mutator phenotype; prophages.
Figures




Similar articles
-
Chromosomal islands of Streptococcus pyogenes and related streptococci: molecular switches for survival and virulence.Front Cell Infect Microbiol. 2014 Aug 12;4:109. doi: 10.3389/fcimb.2014.00109. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25161960 Free PMC article. Review.
-
Phage-associated mutator phenotype in group A streptococcus.J Bacteriol. 2008 Oct;190(19):6290-301. doi: 10.1128/JB.01569-07. Epub 2008 Aug 1. J Bacteriol. 2008. PMID: 18676670 Free PMC article.
-
Genome analysis of an inducible prophage and prophage remnants integrated in the Streptococcus pyogenes strain SF370.Virology. 2002 Oct 25;302(2):245-58. doi: 10.1006/viro.2002.1570. Virology. 2002. PMID: 12441069
-
Elimination of Chromosomal Island SpyCIM1 from Streptococcus pyogenes Strain SF370 Reverses the Mutator Phenotype and Alters Global Transcription.PLoS One. 2015 Dec 23;10(12):e0145884. doi: 10.1371/journal.pone.0145884. eCollection 2015. PLoS One. 2015. PMID: 26701803 Free PMC article.
-
The Bacteriophages of Streptococcus pyogenes.Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.gpp3-0059-2018. doi: 10.1128/microbiolspec.GPP3-0059-2018. Microbiol Spectr. 2019. PMID: 31111820 Free PMC article. Review.
Cited by
-
ϕSa3mw Prophage as a Molecular Regulatory Switch of Staphylococcus aureus β-Toxin Production.J Bacteriol. 2019 Jun 21;201(14):e00766-18. doi: 10.1128/JB.00766-18. Print 2019 Jul 15. J Bacteriol. 2019. PMID: 30962356 Free PMC article.
-
Regulatory gene mutation: a driving force behind group a Streptococcus strain- and serotype-specific variation.Mol Microbiol. 2017 Feb;103(4):576-589. doi: 10.1111/mmi.13584. Epub 2016 Dec 19. Mol Microbiol. 2017. PMID: 27868255 Free PMC article. Review.
-
Chromosomal islands of Streptococcus pyogenes and related streptococci: molecular switches for survival and virulence.Front Cell Infect Microbiol. 2014 Aug 12;4:109. doi: 10.3389/fcimb.2014.00109. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25161960 Free PMC article. Review.
-
The Life Cycle Transitions of Temperate Phages: Regulating Factors and Potential Ecological Implications.Viruses. 2022 Aug 28;14(9):1904. doi: 10.3390/v14091904. Viruses. 2022. PMID: 36146712 Free PMC article. Review.
-
A phage satellite tunes inducing phage gene expression using a domesticated endonuclease to balance inhibition and virion hijacking.Nucleic Acids Res. 2021 May 7;49(8):4386-4401. doi: 10.1093/nar/gkab207. Nucleic Acids Res. 2021. PMID: 33823541 Free PMC article.
References
-
- Banks D. J., Porcella S. F., Barbian K. D., Beres S. B., Philips L. E., Voyich J. M., DeLeo F. R., Martin J. M., Somerville G. A., Musser J. M. (2004). Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 190, 727–73810.1086/422697 - DOI - PubMed
-
- Beres S. B., Richter E. W., Nagiec M. J., Sumby P., Porcella S. F., Deleo F. R., Musser J. M. (2006). Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc. Natl. Acad. Sci. U.S.A. 103, 7059–706410.1073/pnas.0510279103 - DOI - PMC - PubMed
-
- Beres S. B., Sylva G. L., Barbian K. D., Lei B., Hoff J. S., Mammarella N. D., Liu M. Y., Smoot J. C., Porcella S. F., Parkins L. D., Campbell D. S., Smith T. M., McCormick J. K., Leung D. Y., Schlievert P. M., Musser J. M. (2002). Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. U.S.A. 99, 10078–1008310.1073/pnas.152298499 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

