Using the Yeast Three-Hybrid System to Identify Proteins that Interact with a Phloem-Mobile mRNA
- PMID: 22969782
- PMCID: PMC3427875
- DOI: 10.3389/fpls.2012.00189
Using the Yeast Three-Hybrid System to Identify Proteins that Interact with a Phloem-Mobile mRNA
Abstract
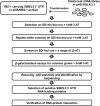
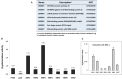
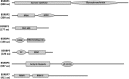
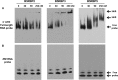
Heterografting and RNA transport experiments have demonstrated the long-distance mobility of StBEL5 RNA, its role in controlling tuber formation, and the function of the 503-nt 3' untranslated region (UTR) of the RNA in mediating transport. Because the 3' UTR of StBEL5 is a key element in regulating several aspects of RNA metabolism, a potato leaf cDNA library was screened using the 3' UTR of StBEL5 as bait in the yeast three-hybrid (Y3H) system to identify putative partner RNA-binding proteins (RBPs). From this screen, 116 positive cDNA clones were isolated based on nutrient selection, HIS3 activation, and lacZ induction and were sequenced and classified. Thirty-five proteins that were predicted to function in either RNA- or DNA-binding were selected from this pool. Seven were monitored for their expression profiles and further evaluated for their capacity to bind to the 3' UTR of StBEL5 using β-galactosidase assays in the Y3H system and RNA gel-shift assays. Among the final selections were two RBPs, a zinc finger protein, and one protein, StLSH10, from a family involved in light signaling. In this study, the Y3H system is presented as a valuable tool to screen and verify interactions between target RNAs and putative RBPs. These results can shed light on the dynamics and composition of plant RNA-protein complexes that function to regulate RNA metabolism.
Keywords: BEL1 family; Solanum tuberosum; StBEL5; mobile RNA; potato; yeast three-hybrid.
Figures



Similar articles
-
Untranslated regions of a mobile transcript mediate RNA metabolism.Plant Physiol. 2009 Dec;151(4):1831-43. doi: 10.1104/pp.109.144428. Epub 2009 Sep 25. Plant Physiol. 2009. PMID: 19783647 Free PMC article.
-
A model system of development regulated by the long-distance transport of mRNA.J Integr Plant Biol. 2010 Jan;52(1):40-52. doi: 10.1111/j.1744-7909.2010.00911.x. J Integr Plant Biol. 2010. PMID: 20074139 Review.
-
The impact of the long-distance transport of a BEL1-like messenger RNA on development.Plant Physiol. 2013 Feb;161(2):760-72. doi: 10.1104/pp.112.209429. Epub 2012 Dec 6. Plant Physiol. 2013. PMID: 23221774 Free PMC article.
-
Conservation of polypyrimidine tract binding proteins and their putative target RNAs in several storage root crops.BMC Genomics. 2018 Feb 7;19(1):124. doi: 10.1186/s12864-018-4502-7. BMC Genomics. 2018. PMID: 29415650 Free PMC article.
-
Multiple Mobile mRNA Signals Regulate Tuber Development in Potato.Plants (Basel). 2017 Feb 10;6(1):8. doi: 10.3390/plants6010008. Plants (Basel). 2017. PMID: 28208608 Free PMC article. Review.
Cited by
-
Phloem RNA-binding proteins as potential components of the long-distance RNA transport system.Front Plant Sci. 2013 May 10;4:130. doi: 10.3389/fpls.2013.00130. eCollection 2013. Front Plant Sci. 2013. PMID: 23675378 Free PMC article.
-
BEL transcription factors in prominent Solanaceae crops: the missing pieces of the jigsaw in plant development.Planta. 2023 Dec 9;259(1):14. doi: 10.1007/s00425-023-04289-8. Planta. 2023. PMID: 38070043 Review.
-
The mRNA mobileome: challenges and opportunities for deciphering signals from the noise.Plant Cell. 2023 May 29;35(6):1817-1833. doi: 10.1093/plcell/koad063. Plant Cell. 2023. PMID: 36881847 Free PMC article. Review.
-
A perspective on photoperiodic phloem-mobile signals that control development.Front Plant Sci. 2013 Aug 2;4:295. doi: 10.3389/fpls.2013.00295. eCollection 2013. Front Plant Sci. 2013. PMID: 23935603 Free PMC article.
-
Protein Interactomics by Two-Hybrid Methods.Methods Mol Biol. 2018;1794:1-14. doi: 10.1007/978-1-4939-7871-7_1. Methods Mol Biol. 2018. PMID: 29855947 Free PMC article.
References
-
- Baroja-Fernandez E., Munoz F. J., Montero M., Etxeberria E., Sesma M. T., Ovecka M., Bahaji A., Ezquer I., Li J., Prat S., Pozueta-Romero J. (2009). Enhancing sucrose synthase activity in transgenic potato (Solanum tuberosum L.) tubers results in increased levels of starch, ADPglucose and UDPglucose and total yield. Plant Cell Physiol. 50, 1651–166210.1093/pcp/pcp108 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

