Development of a survival prediction model for gastric cancer using serine proteases and their inhibitors
- PMID: 22969854
- PMCID: PMC3438548
- DOI: 10.3892/etm.2011.353
Development of a survival prediction model for gastric cancer using serine proteases and their inhibitors
Abstract
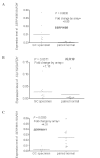
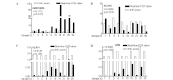
Proteolytic enzymes play a key role in the metastatic stage of gastric cancer (GC). In this study, we aimed to identify the serine proteases (SPs) and their inhibitors (serpins) as related to GC. The gene expression profiles of 40 cases of GC were initially detected by cDNA microarray. The results of the differentially expressed SPs and their inhibitor genes from the microarrays were confirmed by real-time PCR. The status of the immunohistochemical staining of the confirmed genes in patients with complete data was used to develop a survival prediction model. Finally, the prediction model was tested in different groups of GC patients. As a result, seven genes, SERPINB5, KLK10, KLK11, HPN, SPINK1, SERPINA5 and PRSS8, were considered as GC progression-related genes. A survival prediction model including the immunohistochemical scores of three genes and the tumor node metastasis (TNM) score was developed: Survival time (months) = 88.8607 + 2.6395 SERPINB5 - 12.0772 KLK10 + 13.7562 KLK11 - 7.0318 TNM. In conclusion, SERPINB5, KLK10, KLK11, HPN, SPINK1, SERPINA5 and PRSS8 were GC progression-related SPs or serpin genes. The model consisting of the expression profiles of three genes extracted from the microarray study accompanied by the TNM score accurately predicts surgery-related survival of GC patients.
Figures


Similar articles
-
Elevated tumor tissue protein expression levels of kallikrein-related peptidases KLK10 and KLK11 are associated with a better prognosis in advanced high-grade serous ovarian cancer patients.Am J Cancer Res. 2018 Sep 1;8(9):1856-1864. eCollection 2018. Am J Cancer Res. 2018. PMID: 30323977 Free PMC article.
-
Differential gene expression of serine protease inhibitors in bovine ovarian follicle: possible involvement in follicular growth and atresia.Reprod Biol Endocrinol. 2011 May 27;9:72. doi: 10.1186/1477-7827-9-72. Reprod Biol Endocrinol. 2011. PMID: 21619581 Free PMC article.
-
Clinical significance of human kallikrein 10 gene expression in colorectal cancer and gastric cancer.J Gastroenterol Hepatol. 2006 Oct;21(10):1596-603. doi: 10.1111/j.1440-1746.2006.04228.x. J Gastroenterol Hepatol. 2006. PMID: 16928223
-
SerpinB5 interacts with KHDRBS3 and FBXO32 in gastric cancer cells.Oncol Rep. 2011 Nov;26(5):1115-20. doi: 10.3892/or.2011.1369. Epub 2011 Jul 1. Oncol Rep. 2011. PMID: 21725612
-
Clinical and prognostic significances of cancer stem cell markers in gastric cancer patients: a systematic review and meta-analysis.Cancer Cell Int. 2021 Feb 27;21(1):139. doi: 10.1186/s12935-021-01840-z. Cancer Cell Int. 2021. PMID: 33639931 Free PMC article. Review.
Cited by
-
SERPINA5 promotes tumour cell proliferation by modulating the PI3K/AKT/mTOR signalling pathway in gastric cancer.J Cell Mol Med. 2022 Sep;26(18):4837-4846. doi: 10.1111/jcmm.17514. Epub 2022 Aug 24. J Cell Mol Med. 2022. PMID: 36000536 Free PMC article.
-
Discovery and validation of an expression signature for recurrence prediction in high-risk diffuse-type gastric cancer.Gastric Cancer. 2021 May;24(3):655-665. doi: 10.1007/s10120-021-01155-y. Epub 2021 Feb 1. Gastric Cancer. 2021. PMID: 33523340
-
Design and Development of an Intelligent System for Predicting 5-Year Survival in Gastric Cancer.Clin Med Insights Oncol. 2022 Aug 22;16:11795549221116833. doi: 10.1177/11795549221116833. eCollection 2022. Clin Med Insights Oncol. 2022. PMID: 36035639 Free PMC article.
-
Knockdown of KLK11 reverses oxaliplatin resistance by inhibiting proliferation and activating apoptosis via suppressing the PI3K/AKT signal pathway in colorectal cancer cell.Onco Targets Ther. 2018 Feb 16;11:809-821. doi: 10.2147/OTT.S151867. eCollection 2018. Onco Targets Ther. 2018. PMID: 29497313 Free PMC article.
-
Urinary kallikrein 10 predicts the incurability of gastric cancer.Oncotarget. 2017 Apr 25;8(17):29247-29257. doi: 10.18632/oncotarget.16453. Oncotarget. 2017. PMID: 28418926 Free PMC article.
References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
-
- Chau I, Norman AR, Cunningham D, Waters JS, Oates J, Ross PJ. Multivariate prognostic factor analysis in locally advanced and metastatic esophago-gastric cancer – pooled analysis from three multicenter, randomized, controlled trials using individual patient data. J Clin Oncol. 2004;22:2395–2403. - PubMed
-
- Hohenberger P, Gretschel S. Gastric cancer. Lancet. 2003;362:305–315. - PubMed
-
- Curran S, Murray GI. Matrix metalloproteinases in tumour invasion and metastasis. J Pathol. 1999;189:300–308. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
