Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts
- PMID: 22970118
- PMCID: PMC3435383
- DOI: 10.1371/journal.pone.0042786
Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts
Abstract
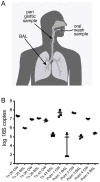
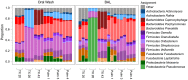
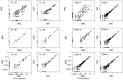
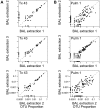
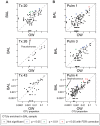
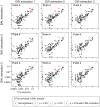
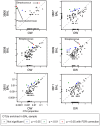
Microbes of the human respiratory tract are important in health and disease, but accurate sampling of the lung presents challenges. Lung microbes are commonly sampled by bronchoscopy, but to acquire samples the bronchoscope must pass through the upper respiratory tract, which is rich in microbes. Here we present methods to identify authentic lung microbiota in bronchoalveolar lavage (BAL) fluid that contains substantial oropharyngeal admixture. We studied clinical BAL samples from six selected subjects with potential heavy lung colonization. A single sample of BAL fluid was obtained from each subject along with contemporaneous oral wash (OW) to sample the oropharynx, and then DNA was extracted from three separate aliquots of each. Bacterial 16S rDNA sequences were amplified and products analyzed by 454 pyrosequencing. By comparing replicates, we were able to specify the depth of sequencing needed to reach a 95% chance of identifying a bacterial lineage of a given proportion--for example, at a depth of 5,000 tags, OTUs of proportion 0.3% or greater would be called with 95% confidence. We next constructed a single-sided outlier test that allowed lung-enriched organisms to be quantified against a background of oropharyngeal admixture, and assessed improvements available with replicate sequence analysis. This allowed identification of lineages enriched in lung in some BAL specimens. Finally, using samples from healthy volunteers collected at multiple sites in the upper respiratory tract, we show that OW provides a reasonable but not perfect surrogate for bacteria carried into to the lung by a bronchoscope. These methods allow identification of microbes that can replicate in the lung despite the background due to oropharyngeal microbes derived from aspiration and bronchoscopic carry-over.
Conflict of interest statement
Figures

References
-
- Jordan GW, Wong GA, Hoeprich PD (1976) Bacteriology of the lower respiratory tract as determined by fiber-optic bronchoscopy and transtracheal aspiration. J Infect Dis 134: 428–435. - PubMed
-
- Kirkpatrick MB, Bass JB Jr (1989) Quantitative bacterial cultures of bronchoalveolar lavage fluids and protected brush catheter specimens from normal subjects. Am Rev Respir Dis 139: 546–548. - PubMed
-
- Pecora DV (1963) A Comparison of Transtracheal Aspiration with Other Methods of Determining the Bacterial Flora of the Lower Respiratory Tract. N Engl J Med 269: 664–666. - PubMed
-
- Bartlett JG, Alexander J, Mayhew J, Sullivan-Sigler N, Gorbach SL (1976) Should fiberoptic bronchoscopy aspirates be cultured? Am Rev Respir Dis 114: 73–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

