Automated reconstruction algorithm for identification of 3D architectures of cribriform ductal carcinoma in situ
- PMID: 22970156
- PMCID: PMC3435324
- DOI: 10.1371/journal.pone.0044011
Automated reconstruction algorithm for identification of 3D architectures of cribriform ductal carcinoma in situ
Abstract
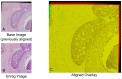
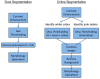
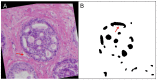
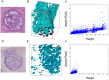
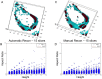
Ductal carcinoma in situ (DCIS) is a pre-invasive carcinoma of the breast that exhibits several distinct morphologies but the link between morphology and patient outcome is not clear. We hypothesize that different mechanisms of growth may still result in similar 2D morphologies, which may look different in 3D. To elucidate the connection between growth and 3D morphology, we reconstruct the 3D architecture of cribriform DCIS from resected patient material. We produce a fully automated algorithm that aligns, segments, and reconstructs 3D architectures from microscopy images of 2D serial sections from human specimens. The alignment algorithm is based on normalized cross correlation, the segmentation algorithm uses histogram equilization, Otsu's thresholding, and morphology techniques to segment the duct and cribra. The reconstruction method combines these images in 3D. We show that two distinct 3D architectures are indeed found in samples whose 2D histological sections are similarly identified as cribriform DCIS. These differences in architecture support the hypothesis that luminal spaces may form due to different mechanisms, either isolated cell death or merging fronds, leading to the different architectures. We find that out of 15 samples, 6 were found to have 'bubble-like' cribra, 6 were found to have 'tube-like' criba and 3 were 'unknown.' We propose that the 3D architectures found, 'bubbles' and 'tubes', account for some of the heterogeneity of the disease and may be prognostic indicators of different patient outcomes.
Conflict of interest statement
Figures


References
-
- Cornfield DB, Palazzo JP, Schwartz GF, Goonewardene SA, Kovatich AJ, et al. (2004) The prognostic significance of multiple morphologic features and biologic markers in ductal carcinoma in situ of the breast: a study of a large cohort of patients treated with surgery alone. Cancer 100: 2317–2327. - PubMed
-
- Wiechmann L, Kuerer HM (2008) The molecular journey from ductal carcinoma in situ to invasive breast cancer. Cancer 112: 2130–2142. - PubMed
-
- Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, et al. (1998) Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci U S A 95: 14821–14826. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

