Pseudomonas aeruginosa exhibits frequent recombination, but only a limited association between genotype and ecological setting
- PMID: 22970178
- PMCID: PMC3435406
- DOI: 10.1371/journal.pone.0044199
Pseudomonas aeruginosa exhibits frequent recombination, but only a limited association between genotype and ecological setting
Abstract
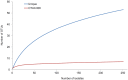
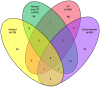
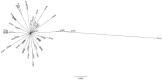
Pseudomonas aeruginosa is an opportunistic pathogen and an important cause of infection, particularly amongst cystic fibrosis (CF) patients. While specific strains capable of patient-to-patient transmission are known, many infections appear to be caused by unique and unrelated strains. There is a need to understand the relationship between strains capable of colonising the CF lung and the broader set of P. aeruginosa isolates found in natural environments. Here we report the results of a multilocus sequence typing (MLST)-based study designed to understand the genetic diversity and population structure of an extensive regional sample of P. aeruginosa isolates from South East Queensland, Australia. The analysis is based on 501 P. aeruginosa isolates obtained from environmental, animal and human (CF and non-CF) sources with particular emphasis on isolates from the Lower Brisbane River and isolates from CF patients obtained from the same geographical region. Overall, MLST identified 274 different sequence types, of which 53 were shared between one or more ecological settings. Our analysis revealed a limited association between genotype and environment and evidence of frequent recombination. We also found that genetic diversity of P. aeruginosa in Queensland, Australia was indistinguishable from that of the global P. aeruginosa population. Several CF strains were encountered frequently in multiple ecological settings; however, the most frequently encountered CF strains were confined to CF patients. Overall, our data confirm a non-clonal epidemic structure and indicate that most CF strains are a random sample of the broader P. aeruginosa population. The increased abundance of some CF strains in different geographical regions is a likely product of chance colonisation events followed by adaptation to the CF lung and horizontal transmission among patients.
Conflict of interest statement
Figures




References
-
- Levin BR, Lipsitch M, Bonhoeffer S (1999) Population biology, evolution, and infectious disease: convergence and synthesis. Science 283: 806–809. - PubMed
-
- Guttman DS, Stavrinides J (2010) Population Genomics of Bacteria. In: Robinson DA, Falush D, Feil EJ, editors. Bacterial Population Genetics in Infectious Disease. Hoboken, New Jersey: John Wiley & Sons, Inc. 121–151.
-
- Westman EL, Matewish M, Lam JS (2010) Pseudomonas. In: Gyles CL, Prescott JF, Songer JG, Thoen CO, editors. Pathogenesis of Bacterial Infections in Animals. Fourth ed. Hoboken, NJ, USA: Wiley-Blackwell. 443–468.
-
- Palleroni NJ (2008) The road to the taxonomy of Pseudomonas. In: Cornelis P, editor. Pseudomonas: Genomic and Molecular Biology. Norfolk: Caister Academic Press. 1–18.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

