Monitoring CSF proteome alterations in amyotrophic lateral sclerosis: obstacles and perspectives in translating a novel marker panel to the clinic
- PMID: 22970211
- PMCID: PMC3435306
- DOI: 10.1371/journal.pone.0044401
Monitoring CSF proteome alterations in amyotrophic lateral sclerosis: obstacles and perspectives in translating a novel marker panel to the clinic
Abstract
Background: Amyotrophic lateral sclerosis (ALS) is a fatal disorder of the motor neuron system with poor prognosis and marginal therapeutic options. Current clinical diagnostic criteria are based on electrophysiological examination and exclusion of other ALS-mimicking conditions. Neuroprotective treatments are, however, most promising in early disease stages. Identification of disease-specific CSF biomarkers and associated biochemical pathways is therefore most relevant to monitor disease progression, response to neuroprotective agents and to enable early inclusion of patients into clinical trials.
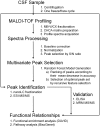
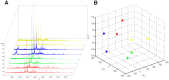
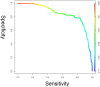
Methods and findings: CSF from 35 patients with ALS diagnosed according to the revised El Escorial criteria and 23 age-matched controls was processed using paramagnetic bead chromatography for protein isolation and subsequently analyzed by MALDI-TOF mass spectrometry. CSF protein profiles were integrated into a Random Forest model constructed from 153 mass peaks. After reducing this peak set to the top 25%, a classifier was built which enabled prediction of ALS with high accuracy, sensitivity and specificity. Further analysis of the identified peptides resulted in a panel of five highly sensitive ALS biomarkers. Upregulation of secreted phosphoprotein 1 in ALS-CSF samples was confirmed by univariate analysis of ELISA and mass spectrometry data. Further quantitative validation of the five biomarkers was achieved in an 80-plex Multiple Reaction Monitoring mass spectrometry assay.
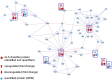
Conclusions: ALS classification based on the CSF biomarker panel proposed in this study could become a valuable predictive tool for early clinical risk stratification. Of the numerous CSF proteins identified, many have putative roles in ALS-related metabolic processes, particularly in chromogranin-mediated secretion signaling pathways. While a stand-alone clinical application of this classifier will only be possible after further validation and a multicenter trial, it could be readily used to complement current ALS diagnostics and might also provide new insights into the pathomechanisms of this disease in the future.
Conflict of interest statement
Figures

References
-
- Bruijn LI, Miller TM, Cleveland DW (2004) Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci 27: 723–749. - PubMed
-
- Pasinelli P, Brown RH (2006) Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci 7: 710–723. - PubMed
-
- Brooks BR, Miller RG, Swash M, Munsat TL (2000) El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 1: 293–299. - PubMed
-
- Traynor BJ, Codd MB, Corr B, Forde C, Frost E, et al. (2000) Clinical features of amyotrophic lateral sclerosis according to the El Escorial and Airlie House diagnostic criteria: A population-based study. Arch Neurol. 57: 1171–1176. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

