Protein disulfide isomerase-like protein 1-1 controls endosperm development through regulation of the amount and composition of seed proteins in rice
- PMID: 22970232
- PMCID: PMC3435311
- DOI: 10.1371/journal.pone.0044493
Protein disulfide isomerase-like protein 1-1 controls endosperm development through regulation of the amount and composition of seed proteins in rice
Abstract
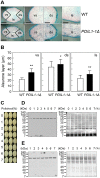
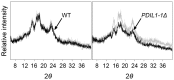
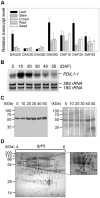
Protein disulfide isomerase (PDI) is a chaperone protein involved in oxidative protein folding by acting as a catalyst and assisting folding in the endoplasmic reticulum (ER). A genome database search showed that rice contains 19 PDI-like genes. However, their functions are not clearly identified. This paper shows possible functions of rice PDI-like protein 1-1 (PDIL1-1) during seed development. Seeds of the T-DNA insertion PDIL1-1 mutant, PDIL1-1Δ, identified by genomic DNA PCR and western blot analysis, display a chalky phenotype and a thick aleurone layer. Protein content per seed was significantly lower and free sugar content higher in PDIL1-1Δ mutant seeds than in the wild type. Proteomic analysis of PDIL1-1Δ mutant seeds showed that PDIL1-1 is post-translationally regulated, and its loss causes accumulation of many types of seed proteins including glucose/starch metabolism- and ROS (reactive oxygen species) scavenging-related proteins. In addition, PDIL1-1 strongly interacts with the cysteine protease OsCP1. Our data indicate that the opaque phenotype of PDIL1-1Δ mutant seeds results from production of irregular starch granules and protein body through loss of regulatory activity for various proteins involved in the synthesis of seed components.
Conflict of interest statement
Figures



References
-
- Tsai C, Larkins B, Glover D (1978) Interaction of the opaque-2 gene with starch-forming mutant genes on the synthesis of zein in maize endosperm. Biochem Genet 16: 883–896. - PubMed
-
- Miles MJ, Morris VJ, Orford PD, Ring SG (1985) The roles of amylose and amylopectin in the gelation and retrogradation of starch. Carbohydr Res 135: 271–281.
-
- Gallant DJ, Bouchet B, Baldwin PM (1997) Microscopy of starch: evidence of a new level of granule organization. Carbohydr Polym 32: 177–191.
-
- Thompson DB (2000) On the non-random nature of amylopectin branching. Carbohydr. Polym 43: 223–239.
-
- Smith AM, Denyer K, Martin C (1997) The synthesis of the starch granule. Ann Rev Plant Biol 48: 67–87. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

