Hypoxic induction of the regulator of G-protein signalling 4 gene is mediated by the hypoxia-inducible factor pathway
- PMID: 22970249
- PMCID: PMC3436875
- DOI: 10.1371/journal.pone.0044564
Hypoxic induction of the regulator of G-protein signalling 4 gene is mediated by the hypoxia-inducible factor pathway
Abstract
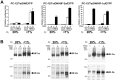
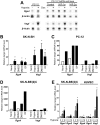
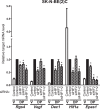
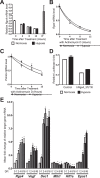
The transcriptional response to hypoxia is largely dependent on the Hypoxia Inducible Factors (HIF-1 and HIF-2) in mammalian cells. Many target genes have been characterised for these heterodimeric transcription factors, yet there is evidence that the full range of HIF-regulated genes has not yet been described. We constructed a TetON overexpression system in the rat pheochromocytoma PC-12 cell line to search for novel HIF and hypoxia responsive genes. The Rgs4 gene encodes the Regulator of G-Protein Signalling 4 (RGS4) protein, an inhibitor of signalling from G-protein coupled receptors, and dysregulation of Rgs4 is linked to disease states such as schizophrenia and cardiomyopathy. Rgs4 was found to be responsive to HIF-2α overexpression, hypoxic treatment, and hypoxia mimetic drugs in PC-12 cells. Similar responses were observed in human neuroblastoma cell lines SK-N-SH and SK-N-BE(2)C, but not in endothelial cells, where Rgs4 transcript is readily detected but does not respond to hypoxia. Furthermore, this regulation was found to be dependent on transcription, and occurs in a manner consistent with direct HIF transactivation of Rgs4 transcription. However, no HIF binding site was detectable within 32 kb of the human Rgs4 gene locus, leading to the possibility of regulation by long-distance genomic interactions. Further research into Rgs4 regulation by hypoxia and HIF may result in better understanding of disease states such as schizophrenia, and also shed light on the other roles of HIF yet to be discovered.
Conflict of interest statement
Figures
Similar articles
-
The specific contribution of hypoxia-inducible factor-2alpha to hypoxic gene expression in vitro is limited and modulated by cell type-specific and exogenous factors.Exp Cell Res. 2008 Jun 10;314(10):2016-27. doi: 10.1016/j.yexcr.2008.03.003. Epub 2008 Mar 18. Exp Cell Res. 2008. PMID: 18420194
-
Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells.Mol Cell Biol. 2006 May;26(9):3514-26. doi: 10.1128/MCB.26.9.3514-3526.2006. Mol Cell Biol. 2006. PMID: 16611993 Free PMC article.
-
HER2 regulates HIF-2α and drives an increased hypoxic response in breast cancer.Breast Cancer Res. 2019 Jan 22;21(1):10. doi: 10.1186/s13058-019-1097-0. Breast Cancer Res. 2019. PMID: 30670058 Free PMC article.
-
Multiplicity of hypoxia-inducible transcription factors and their connection to the circadian clock in the zebrafish.Physiol Biochem Zool. 2015 Mar-Apr;88(2):146-57. doi: 10.1086/679751. Epub 2015 Jan 14. Physiol Biochem Zool. 2015. PMID: 25730270 Review.
-
Carrot and stick: HIF-alpha engages c-Myc in hypoxic adaptation.Cell Death Differ. 2008 Apr;15(4):672-7. doi: 10.1038/sj.cdd.4402302. Epub 2008 Jan 11. Cell Death Differ. 2008. PMID: 18188166 Review.
Cited by
-
Cofilin Inhibition Restores Neuronal Cell Death in Oxygen-Glucose Deprivation Model of Ischemia.Mol Neurobiol. 2016 Mar;53(2):867-878. doi: 10.1007/s12035-014-9056-3. Epub 2014 Dec 20. Mol Neurobiol. 2016. PMID: 25526862 Free PMC article.
-
Conserved N-terminal cysteine dioxygenases transduce responses to hypoxia in animals and plants.Science. 2019 Jul 5;365(6448):65-69. doi: 10.1126/science.aaw0112. Science. 2019. PMID: 31273118 Free PMC article.
-
Transcriptional regulation by hypoxia inducible factors.Crit Rev Biochem Mol Biol. 2014 Jan-Feb;49(1):1-15. doi: 10.3109/10409238.2013.838205. Epub 2013 Oct 7. Crit Rev Biochem Mol Biol. 2014. PMID: 24099156 Free PMC article. Review.
-
Inducible glomerular erythropoietin production in the adult kidney.Kidney Int. 2015 Dec;88(6):1345-1355. doi: 10.1038/ki.2015.274. Epub 2015 Sep 23. Kidney Int. 2015. PMID: 26398496
-
ZNF804A Transcriptional Networks in Differentiating Neurons Derived from Induced Pluripotent Stem Cells of Human Origin.PLoS One. 2015 Apr 23;10(4):e0124597. doi: 10.1371/journal.pone.0124597. eCollection 2015. PLoS One. 2015. PMID: 25905630 Free PMC article.
References
-
- Wenger RH, Stiehl DP, Camenisch G (2005) Integration of oxygen signaling at the consensus HRE. Sci STKE 2005: re12. - PubMed
-
- Wang GL, Semenza GL (1995) Purification and characterization of hypoxia-inducible factor 1. J Biol Chem 270: 1230–1237. - PubMed
-
- Konietzny R, Konig A, Wotzlaw C, Bernadini A, Berchner-Pfannschmidt U, et al. (2009) Molecular imaging: into in vivo interaction of HIF-1alpha and HIF-2alpha with ARNT. Ann N Y Acad Sci 1177: 74–81. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

